
This blog is adapted from a talk at an MLOps Learners demo day (slides, video). marimo is free and open source, available on GitHub.
I worked on vector embeddings during my PhD at Stanford. I used Jupyter daily, because it paired code with visuals in an iterative programming environment. But I realized early on that we needed a Python notebook that was more than just a REPL. I spent a lot of time thinking about this, perhaps because I had just finished a stint at Google as an engineer working at Google Brain, where I thought a lot about computer systems and dataflow graphs.
A lot of important work happens in notebooks — research, large scale experimentation, data engineering, and more. But this kind of work should be treated as Python software, and it shouldn’t be done in error-prone JSON scratchpads. By the end of my PhD, it became clear to me that we needed to rethink what a notebook is, from the ground up and from first principles.
Desiderata
When setting out to build marimo, I had three desiderata for it:
- Reproducible: well-specified execution order, while still allowing for interactive execution of cells. Your code and outputs should always be in sync, not unlike a spreadsheet.
- Maintainable: pure Python, so code is versionable and portable.
- Multi-purpose: not just a notebook — share as an interactive web app, execute as a script or pipeline, without jumping through extra hoops.
These desiderata build on each other: maintainable requires reproducible, and multi-purpose requires maintainable and reproducible. In this blog post, I’ll talk about how we designed for these desiderata and lessons we learned along the way.
Reproducible
Every notebook should be computationally reproducible: the variables in memory should be a function of code on the page, only, so that your code and outputs are always consistent.
Hidden state
Computational reproducibility sounds straightforward — its how most regular programs work. But it’s not how traditional notebooks, such as Jupyter with IPyKernel, work. Traditional notebooks let developers execute cells one at a time, in any order, with each cell modifying a mutable workspace (i.e., the variables in memory). This creates hidden state: in traditional notebooks, variable state is a function of code and the user’s execution history.
This hidden state is a huge problem for reproducibility. It’s why when you receive a notebook from a colleague and try to run it from top to bottom, the notebook just breaks or has different results from the outputs saved in the notebook.
Here are two examples of hidden state. In the first example,
the cells were executed out of order, and now the page reports x == 0, even
though in fact x == 1.

In this example, x == 0 in memory, but x is not defined on the page! This
is because the author deleted the cell defining x, but the IPython kernel
kept x in memory. Restarting and running this notebook would yield a totally
different result.
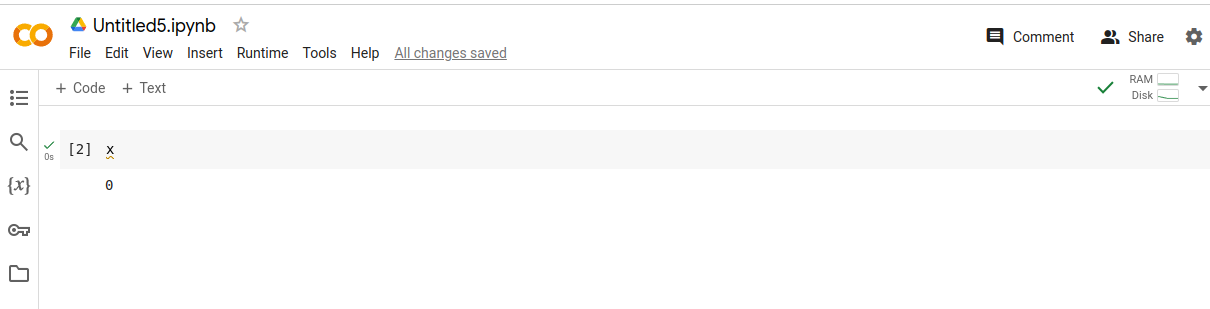
These are toy examples, but what happens in practice is that your co-author or colleague gives you a notebook that does some real task: generates plots for a paper, transforms some data, or runs an experiment, but when you run it, the notebook just doesn’t work the way the author thinks it does. A study of 10 million notebooks on GitHub found that 36% percent of them were executed out of order in this way.
Eliminating hidden state by modeling notebooks as DAGs
To eliminate hidden state, we model every notebook as a directed acyclic graph
(DAG) on cells. We mark each cell with the variables it defines and the
variables it references. There’s an edge (a, b) if b references any of
the variables defined by a. We form this graph using static
analysis, reading your code without running it, so there’s no performance
overhead at runtime.
The DAG encodes dependencies across cells: it specifies how variables flow
from one cell to another. The semantics of the graph are that if a is a parent
of b, meaning that b reads a variable defined by a, then
b has to run after a.
This kind of graph is also known as a dataflow graph. If you’ve used
TensorFlow, TorchScript, JAX — or Excel — you’ve worked with dataflow graphs
before. The nice thing about a dataflow graph is that it imposes a well-defined
execution order on the notebook. If you run a cell that defines a variable y = x + 1, the notebook should:
- first, compute
xif it was defined by another cell that hasn’t run; - then, compute
y = x + 1, ie run the cell; - finally, either automatically run other cells that use
y, or mark them as stale and invalidate their memory.
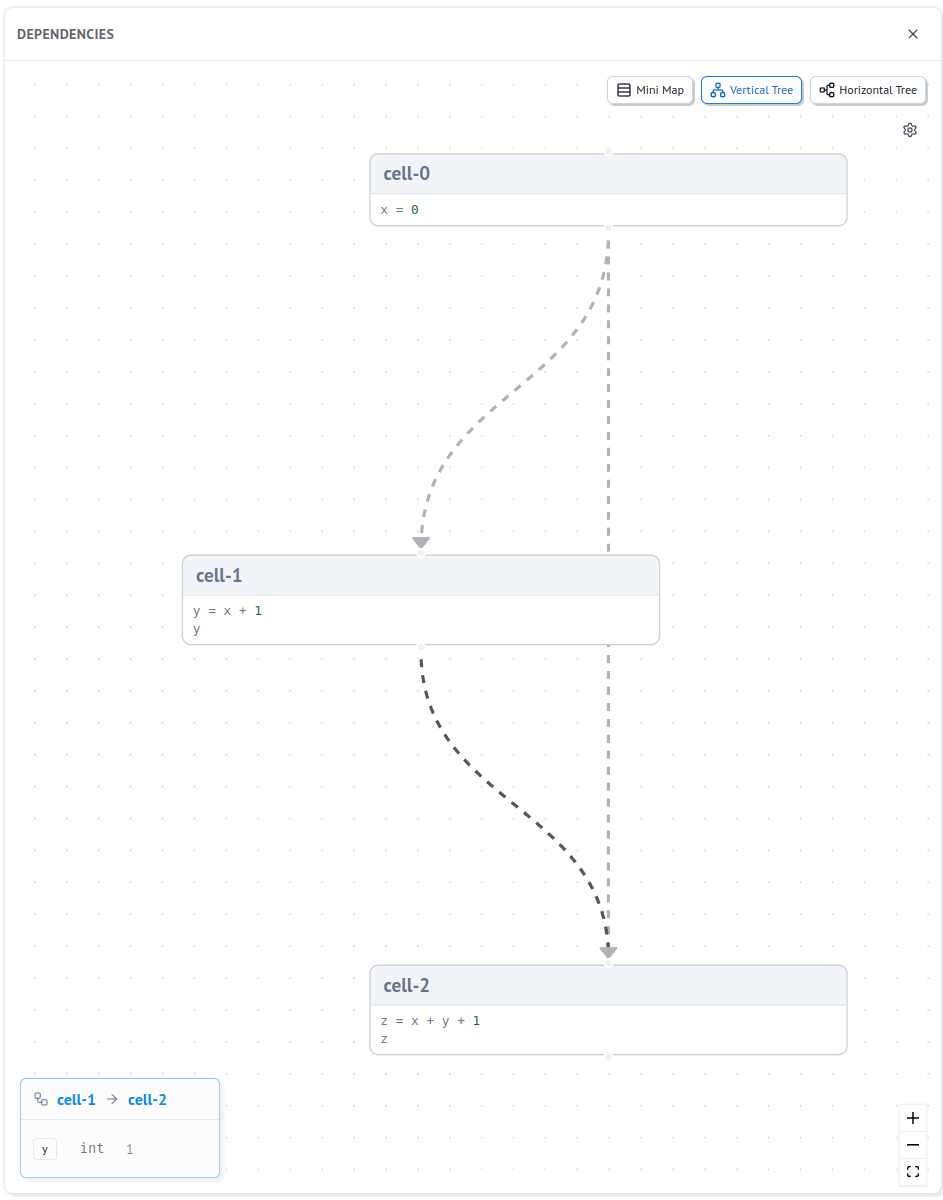
Here is an example marimo notebook for you to play with that makes this concrete.
It has the following dependency graph:
Constraints
To make sure every notebook is a DAG, the user must accept some constraints:
- variables can’t be reassigned
- cells can’t have cycles (
x = y,y = x) - avoid mutating variables across cells (mutations are discouraged but allowed)
These constraints have a learning curve but are easy to understand. And they unlock a lot of new capabilities for the user: not only are DAG-based notebooks reproducible, they can also support interactive UI elements that automatically synchronize with Python:
Execution
By default, marimo automatically updates outputs, like a spreadsheet: when a cell is run, its children are too. But this can be configured to be lazy, so that children are only run when requested and instead marked as stale.
Alternatives considered
We considered a couple alternatives to modeling a notebook as a DAG:
-
Only allow notebooks to be run top-to-bottom? This works for things like streamlit, but is too rigid for interactive computing.
-
Build a dependency graph at runtime? This requires tracing the user code and watching for variable reads/writes. The problem is that we’ll only catch an unspecified subset of reads and writes, so the dependency graph will be ill-defined and users won’t be able to understand the structure of the problem. Plus, tracing adds a substantial performance overhead.
-
Just warn users if they execute things out-of-order? This doesn’t solve the problem and won’t enable the other desiderata, so a non-starter.
Lesson: prefer simplicity
In choosing static inference, we chose a simple way to eliminate hidden state. People are already familiar with dataflow graphs, even if they don’t know it:
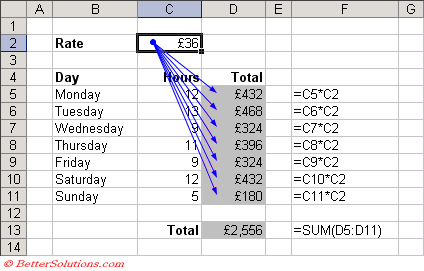 The excel spreadsheet: the original DAG-powered notebook …
The excel spreadsheet: the original DAG-powered notebook …
Yes, there are constraints. But people will happily accept constraints if they are necessary and easily understood.
Other solutions like runtime tracing, or relaxing constraints, were just too complicated for users to think about, let alone me as the developer as it went down the path of solving an unsolvable problem (we learned hard lessons about trying to solve impossible problems at TensorFlow).
My biggest takeaway here is that it’s important to design systems that are easily explained, and constraints are okay. An easily understood system with clear constraints is better than a inscrutable one without constraints.
Maintainable
We grounded maintainability in the file format. Instead of storing notebooks as
JSON, we insisted on storing notebooks as Python files (.py) with the
following properties:
- git-friendly: small code change => small diff
- easy for both humans and computers to read
- importable as a Python module
- executable as a Python script
- editable with a text editor
After a lot of careful consideration, we landed on files that look like this:
import marimo
__generated_with = "0.4.12"
app = marimo.App()
@app.cell
def defines_x():
x = 0
return x,
@app.cell
def defines_y(x):
y = x + 1
y
return y,
@app.cell
def computes_z(x, y):
z = x + y + 1
z
return z,
if __name__ == "__main__":
app.run()Cells are encoded as functions that read their variable references and return
their definitions, and are added to an app object that builds the graph. Users
can optionally name their cells, which become the names of the decorated
functions.
As an alternative, we considered storing files as flat Python scripts,
with comments demarcating cells (instead of putting them in functions). This is
simple and has precedent (VSCode notebooks, Pluto.jl), but it makes importing
notebooks as modules impractical — it either executes all the code on import,
or makes it inaccessible if stored under an if __name__ == "__main__" guard.
Lesson learned: design for extensibility
Our solution is more complicated than a flat script, but it enables composition
— use named cells as functions in other files. It also gives us a reserved
namespace for future APIs, on the app object. We didn’t know what those APIs
would be when we designed the file format, but we did know that we’d have some
APIs, and had to design for that in mind.
Designing for extensibility is hard. But when you’re making something as delicate as a file format — where you want to be both backward and forward compatible — it’s worth getting it right. In our case, we wrote a 2500 word design doc before implementing a single line of code.
Multi-purpose
Our reproducible ✅ and maintainable ✅ notebook is naturally multi-purpose, capable of being used as more than just a notebook.
A multi-purpose notebook
Run as an app. The basis of modern web apps is reactivity: interact with a UI element and the app automatically reacts to the change in its value. Because each notebook is a DAG, and because we have support for UI elements — interact with a slider, and it triggers execution of cells that reference it — we can turn notebooks into apps by just hiding code, showing outputs, and serving in a read-only mode.
Importantly, this requires 0 lines of code changes: just marimo run notebook.py at the CLI to run as a web app.
Execute as a script. In the previous section, we saw that each notebook ends with
if __name__ == "__main__":
app.run()The app.run() line executes the notebook in an order determined by the
DAG. This means you can type
python notebook.pyat the command-line to execute your notebook as a regular script. We have built-in support for command-line arguments, so you can even parametrize notebooks without jumping through extra hoops.
Map over batches of parameters. In principle, we could even re-use notebooks in batch computing settings, by programmatically substituting a variable in the notebook with a batch of variables and executing the DAG with distributed compute. We haven’t built this yet, but it’s just one more example of how modeling notebooks as DAGs unlocks interesting new capabilities.
Lesson: Stay true to your project’s pillars
This is the biggest lesson we’ve learned while working on marimo: stay true to your project’s pillars. This requires figuring out what your pillars are — the core of your project on which everything is built. This can be a technology, a vision, or both.
You should reject feature requests that require you to compromise your pillars, because trying to please everyone usually results in a worse system. But embrace requests that reinforce or build on your pillars. And sometimes, you can learn from the former requests and find a way to meet your users halfway.
Examples
Here’s our main pillar: every notebook is a DAG on blocks of Python code. This single pillar enables many things:
- notebook-style computation and visualization of data, in a reproducible setting;
- running notebooks as interactive web apps;
- executing notebooks as scripts;
- experimenting with batch processing and distributed runtimes.
Here’s a feature request we recently received that tested the strength of our pillar. A user asked us to support Jupyter-style execution — to allow multiple definitions of variables, cycles, and out-of-order execution, i.e., to opt-out of the DAG. In doing so, our user argued that we’d make it easier for Jupyter users to onboard to marimo.
Allowing users to opt-out of the DAG would indeed make marimo more accessible to Jupyter users. But it would also break everything that marimo enables: it would introduce hidden state, make it impossible to run notebooks as apps or execute them as scripts with well-defined execution order, and severely limit the runtimes we could experiment with. For this reason, we politely rejected this feature request.
On the other hand, this user’s feedback stuck with me: they mentioned that automatic execution of cells’ descendants was less than ideal for notebooks with expensive cells. And I think really, this was the heart of their complaint. So we embraced the spirit of their feedback, if not its letter, and added support for a lazy runtime that marks descendants as stale instead of automatically executing them. In this way, we made marimo better suited to running expensive computations, while still retaining the guarantees provided by the underlying DAG.
Another pillar: marimo notebooks are pure Python. If you’ve
been paying close attention, you’ll have noticed that marimo is both a notebook
and a library (import marimo as mo). The library provides access to things
like UI elements, markdown, and other helpful utilities. marimo exists
as a library precisely because we want marimo notebooks to be just Python;
this puts them on the same footing as other Python programs and helps
make them multi-purpose.
That’s why we don’t have support for Jupyter magics. If a user asks for
a magic command, we typically just build the functionality directly
into the notebook (like we did for module reloading),
or suggest a pure Python alternative (e.g., subprocess.run instead of ! commands).
Conclusion
When I first started marimo, I didn’t imagine that it would be used by places like Stanford, BlackRock, CZ Research, SLAC, LBNL, Sonos, Gridmatic, Sumble, and so many others.
We have lots of work ahead of us, and many more lessons to learn. Still, I think the lessons we’ve learned are durable, and I hope they’ll keep us grounded as we grow.
- GitHub: https://github.com/marimo-team/marimo
- Discord: https://marimo.io/discord
- Docs: https://docs.marimo.io
- Newsletter: https://marimo.io/newsletter



