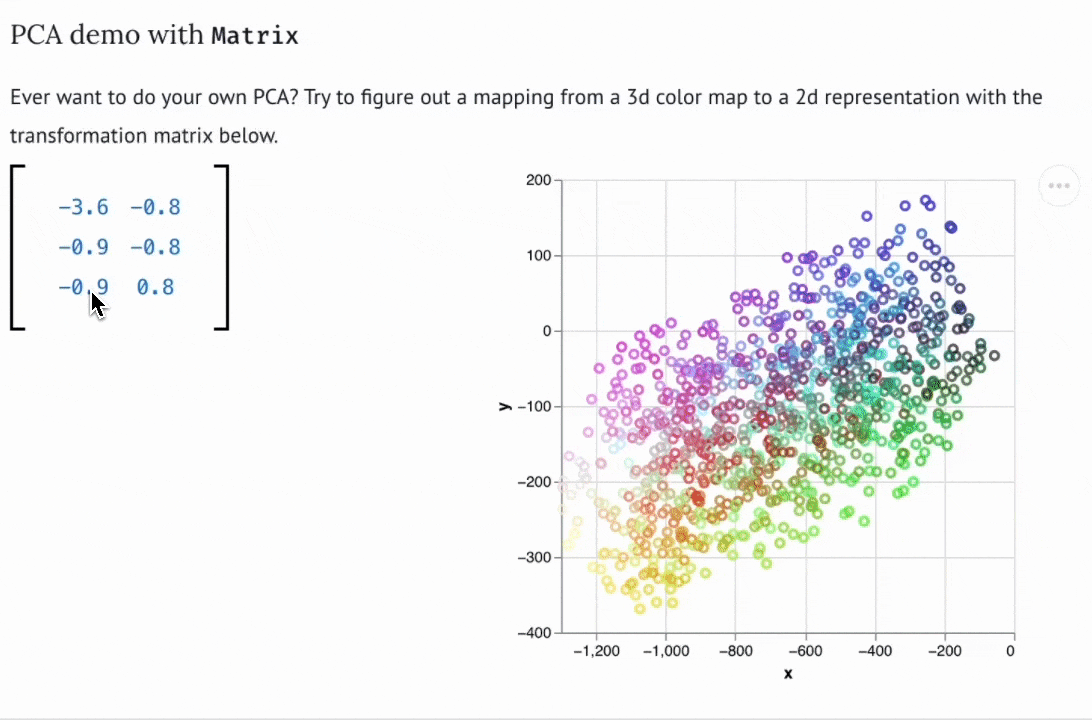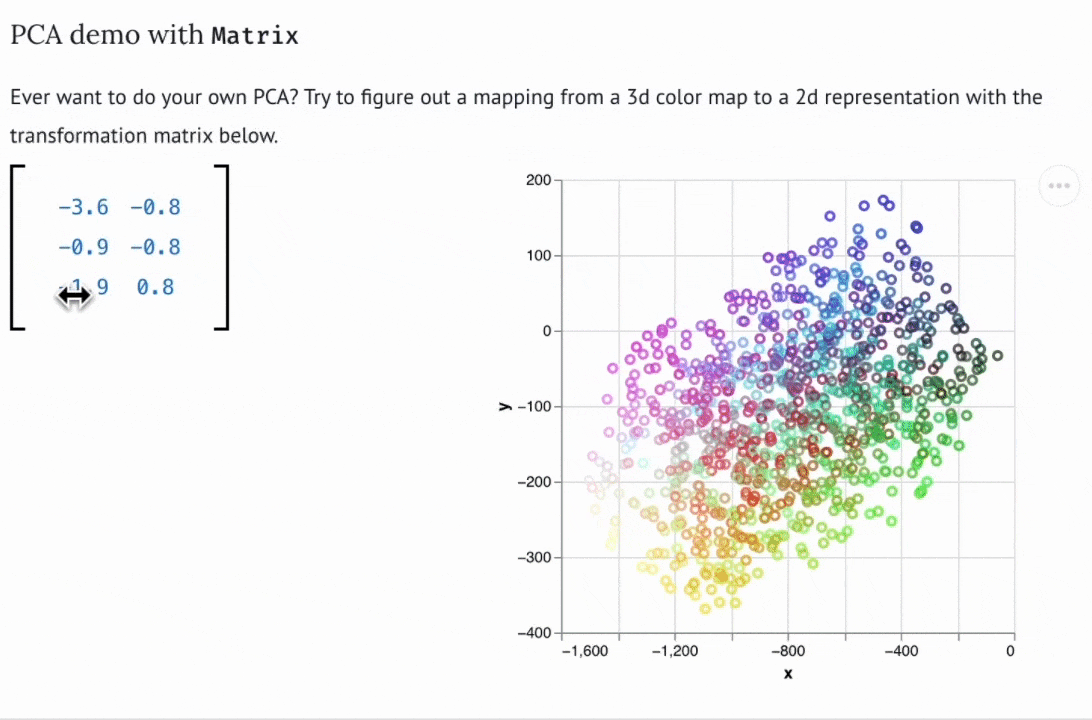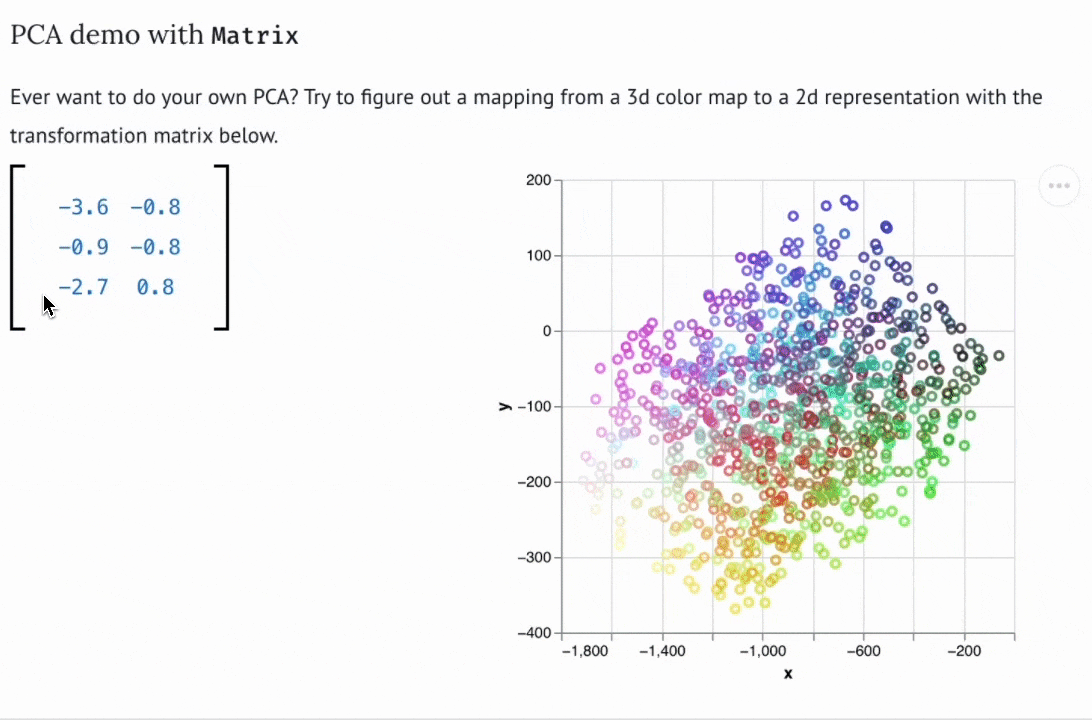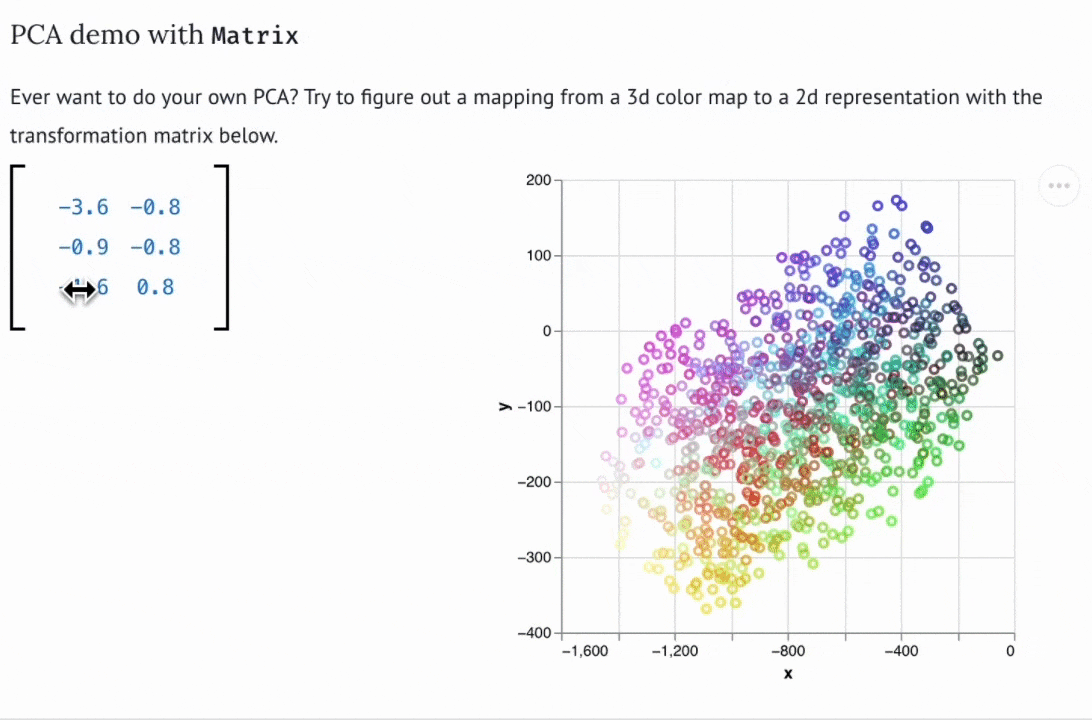
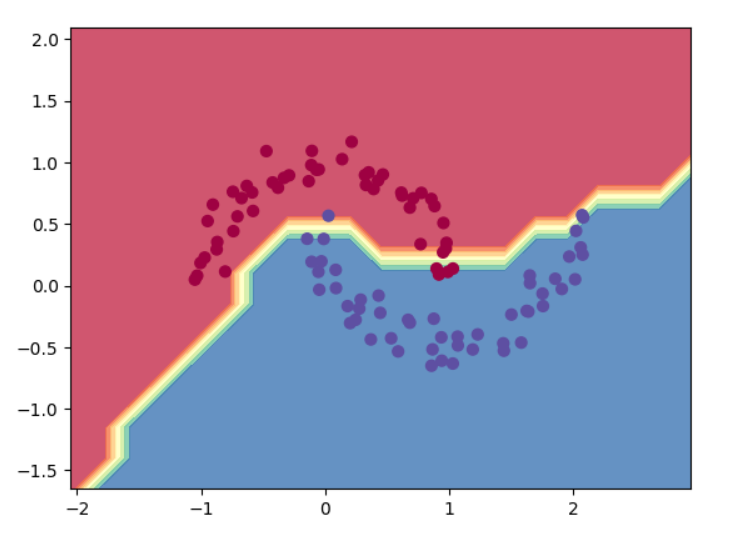
Why Stanford scientists needed a new Python notebook
Bennet Meyers is a staff scientist with Stanford’s SLAC National Accelerator Laboratory; David Chassin is the owner and principal analyst at Eudoxys Sciences, and was previously the group manager, chief scientist, and principal investigator of the GISMo group at SLAC. Akshay and Myles are developers of marimo.
Before it was open source, the marimo Python notebook was originally developed with input from computational scientists at Stanford’s SLAC National Accelerator Laboratory. These scientists needed a new programming environment for their iterative, data-heavy coding work — one that was reproducible and reusable by default.
Jupyter notebooks were historically the workhorse for SLAC’s Grid Integration, Systems, and Mobility (GISMo) group for everything from algorithm development and model validation, to sharing and communicating results. In recent years, however, scientists at GISMo and elsewhere became increasingly aware of limitations with Jupyter that made them unsuited to computational science. Jupyter notebooks suffer from a reproducibility crisis, with less than 4 percent of notebooks on GitHub capable of being reproduced — a problem compounded by how difficult it is to version and test Jupyter notebooks. Moreover, because the final artifact is a static document, with no interactivity, Jupyter’s usefulness for science communication on the web was limited.
These Stanford scientists — authors Bennet Meyers and David Chassin — decided to help shape the development of marimo as a modern alternative to Jupyter, built from the ground up with computational science in mind. Unlike Jupyter notebooks, marimo notebooks are reproducible, stored as Python code, version controllable with Git, shareable as interactive web apps, and executable as scripts.
In this blog post, we describe the design decisions that make marimo well suited to computational science, and how GISMo scientists have used marimo to publish interactive educational lessons, conduct reproducible research using a framework for computational science, and teach students how to analyze data with code.
Making computational notebooks reproducible
The reproducibility crisis affecting Jupyter notebooks is well-documented. In 2019, a study from New York University and Federal Fluminense University found that of the 863,878 Jupyter notebooks on GitHub with valid execution orders, only 24% could be re-run, and just 4% reproduced the same results. A similar study from 2020 by JetBrains found that over a third of the notebooks on GitHub had invalid execution histories. (These problems are specific to the Jupyter notebook’s default experience, i.e. the Jupyter frontends and IPython kernel that nearly all scientists use, not the broader Project Jupyter’s well-thought-out, lower-level protocols.)
Keeping code and outputs in sync with reactive execution
Executing the same notebook should produce the same outputs — like plots and numerical tables, as well as variable values — no matter how the notebook was executed. The marimo notebook was designed with this ideal in mind, but Jupyter notebooks fall far short of it.
Even on the same machine, a scientist can run the same Jupyter notebook twice and produce different results. This shortcoming is intrinsic to the REPL-like experience Jupyter notebooks provide, in which scientists run code cells manually, one-at-a-time, like an Excel spreadsheet in which formulas don’t recalculate. This places an unreasonable cognitive burden on the scientist to maintain the state of their program: if one code cell depends on another, the burden is on the scientist to remember to run the dependent cell; if a cell is deleted, the burden is on the scientist to manually delete its variables from memory and then to re-run all cells that refer to these manually deleted variables. Forget a single step, and your program silently becomes invalid, riddled with hidden state.
In contrast, marimo automatically keeps code and outputs in sync: when the notebook author runs a cell, all other cells that depend on its variables are marked as stale and (optionally) automatically rerun; delete a cell, and its variables are automatically removed from program memory. This “reactive” execution model largely eliminates hidden state, while still letting the author iteratively run parts of the notebook.
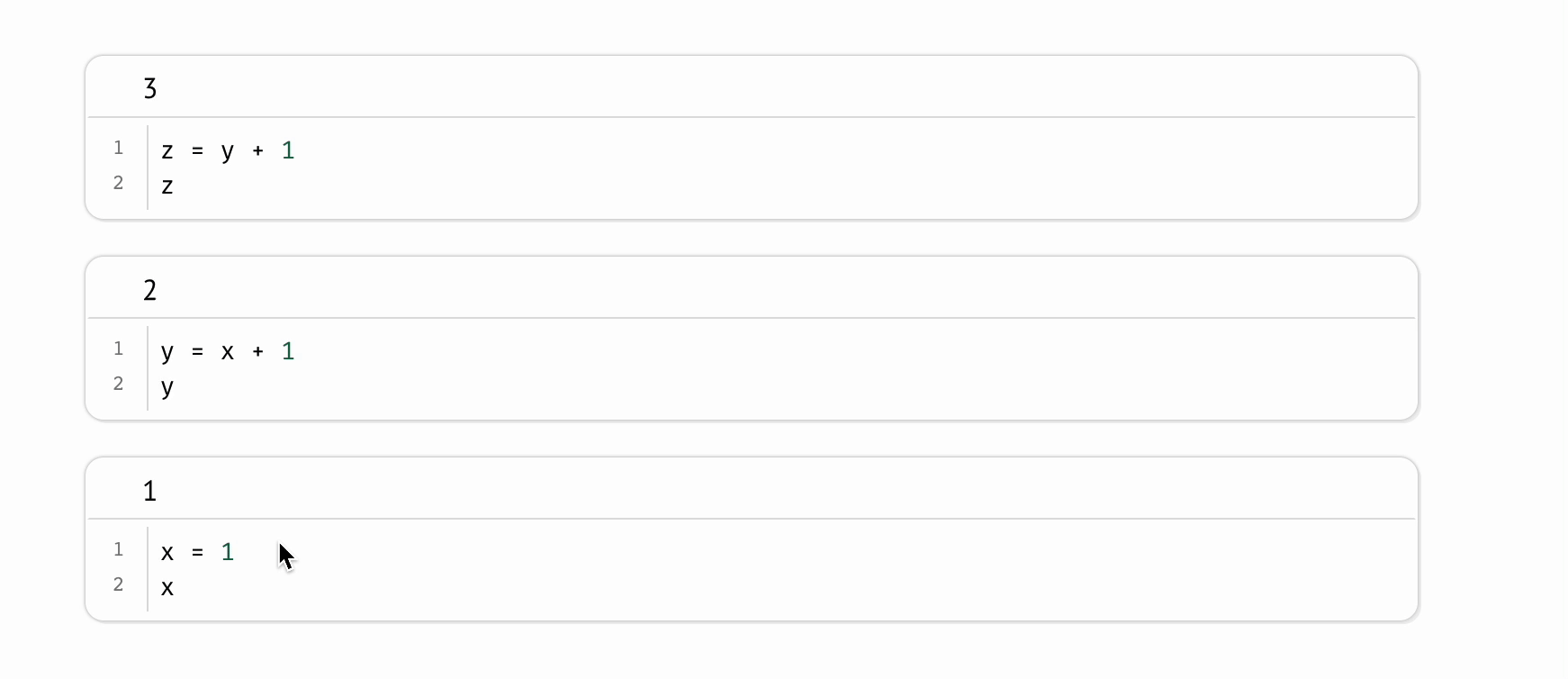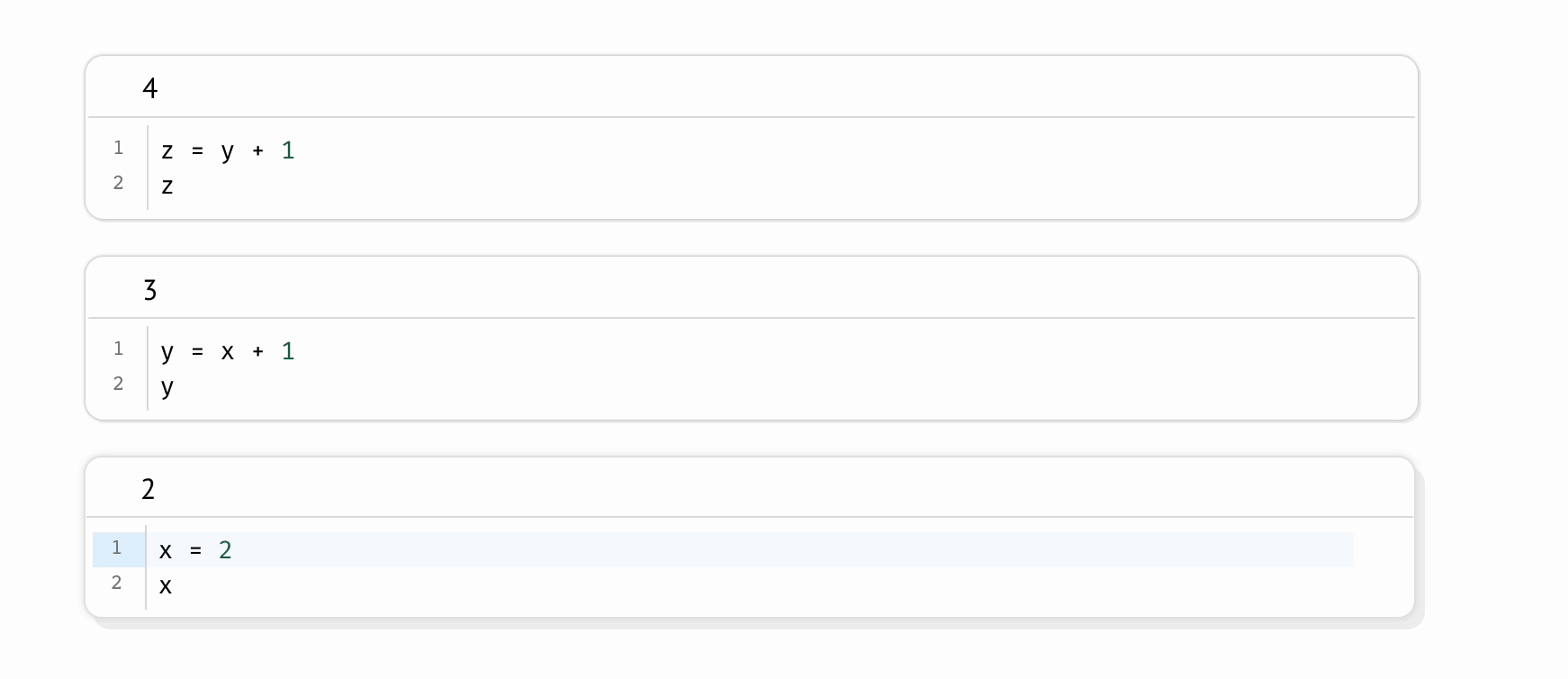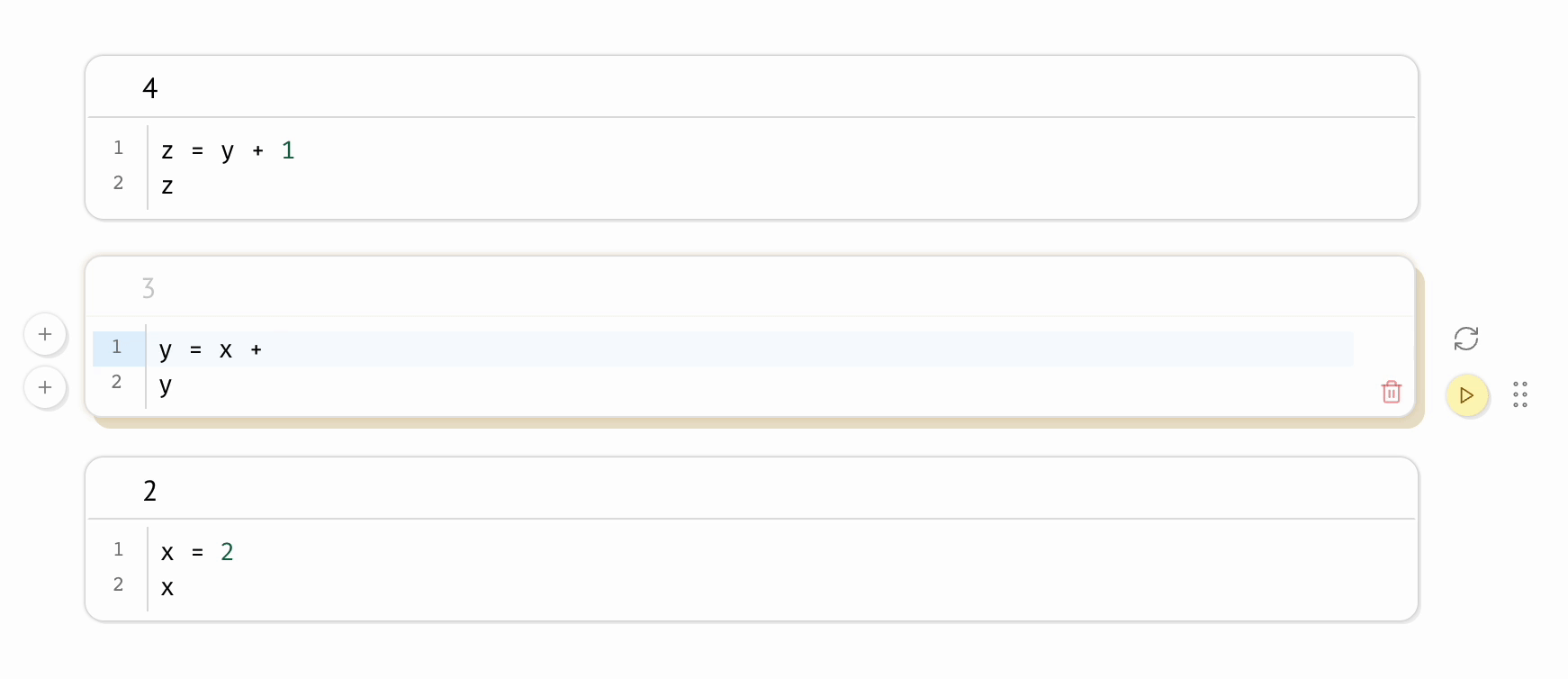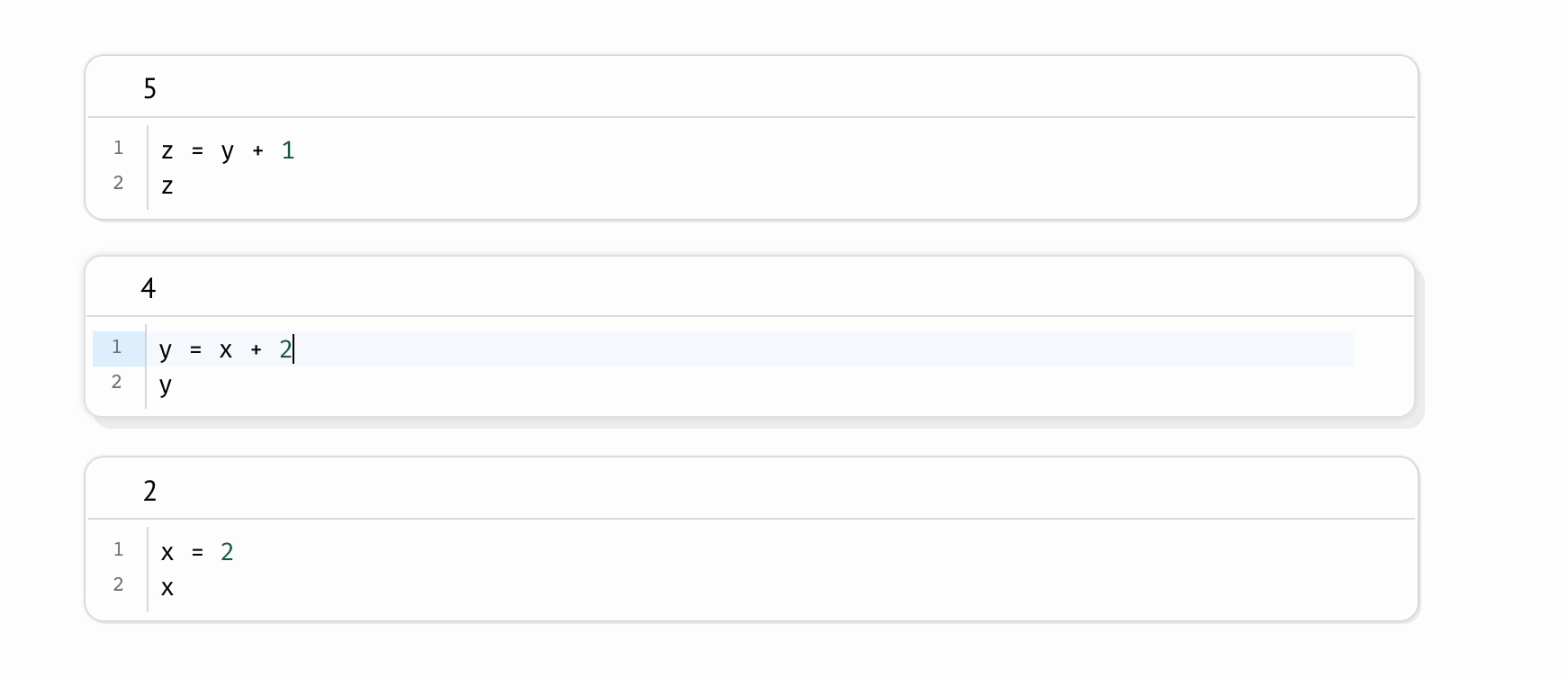
To mitigate the accumulation of hidden state in Jupyter notebooks, it is sometimes recommended to periodically restart notebooks while working on them; but not only does this place yet another burden on the scientist, it is also impractical for expensive notebooks in which computations take significant time or cost money. In contrast, marimo accommodates expensive notebooks by letting developers disable automatic execution while still providing guarantees about state, providing visual cues when a cell’s output becomes outdated.
Reproducible down to the packages
Jupyter notebooks can fail to reproduce because they don’t encapsulate the author’s package environment. For Jupyter, the notebook author has to document the packages used and their versions, as well as provide collaborators with a way to re-instantiate that environment. This is a difficult task in general and it is especially difficult for scientists, who are not typically trained as software engineers.
marimo solves package reproducibility by tracking the package environment in the notebook file itself, similar to Pluto.jl. Given a notebook file, marimo can automatically download its packages into a temporary package environment. This makes it much more likely that a notebook that works on one machine will work an another.
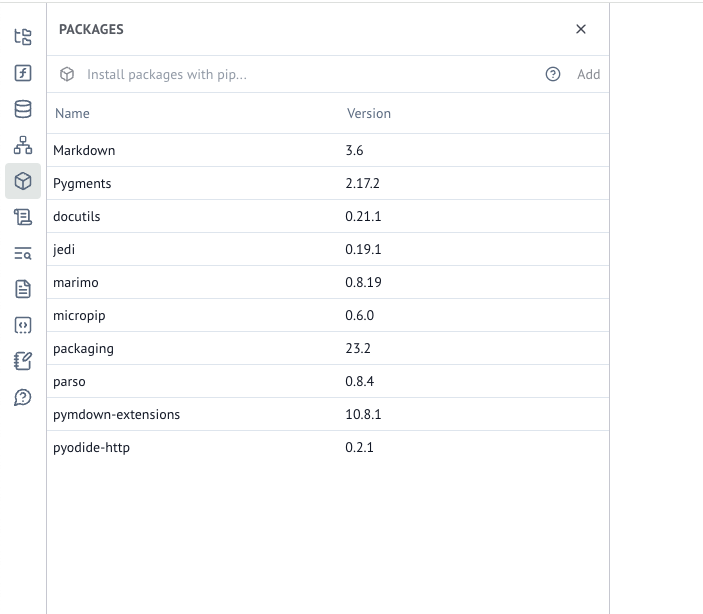
Tracking changes to experiments with version control
Unlike Jupyter notebooks, marimo notebooks are stored as pure Python files, making it easy to use Git to keep track of how an experiment changes over time.
More generally, because marimo notebooks are just Python files, can be reused like any other Python program: execute as Python scripts, test with frameworks like pytest, import into other Python programs, use new package management standards, and more.
Enabling interactive science communication on the web
Reactive execution not only makes notebooks reproducible; it also makes it easy to use interactive widgets. marimo provides a large library of widgets out of the box, like siders, dropdown menus, and selectable charts, to allow both scientists and readers to explore how changing variables affects results.
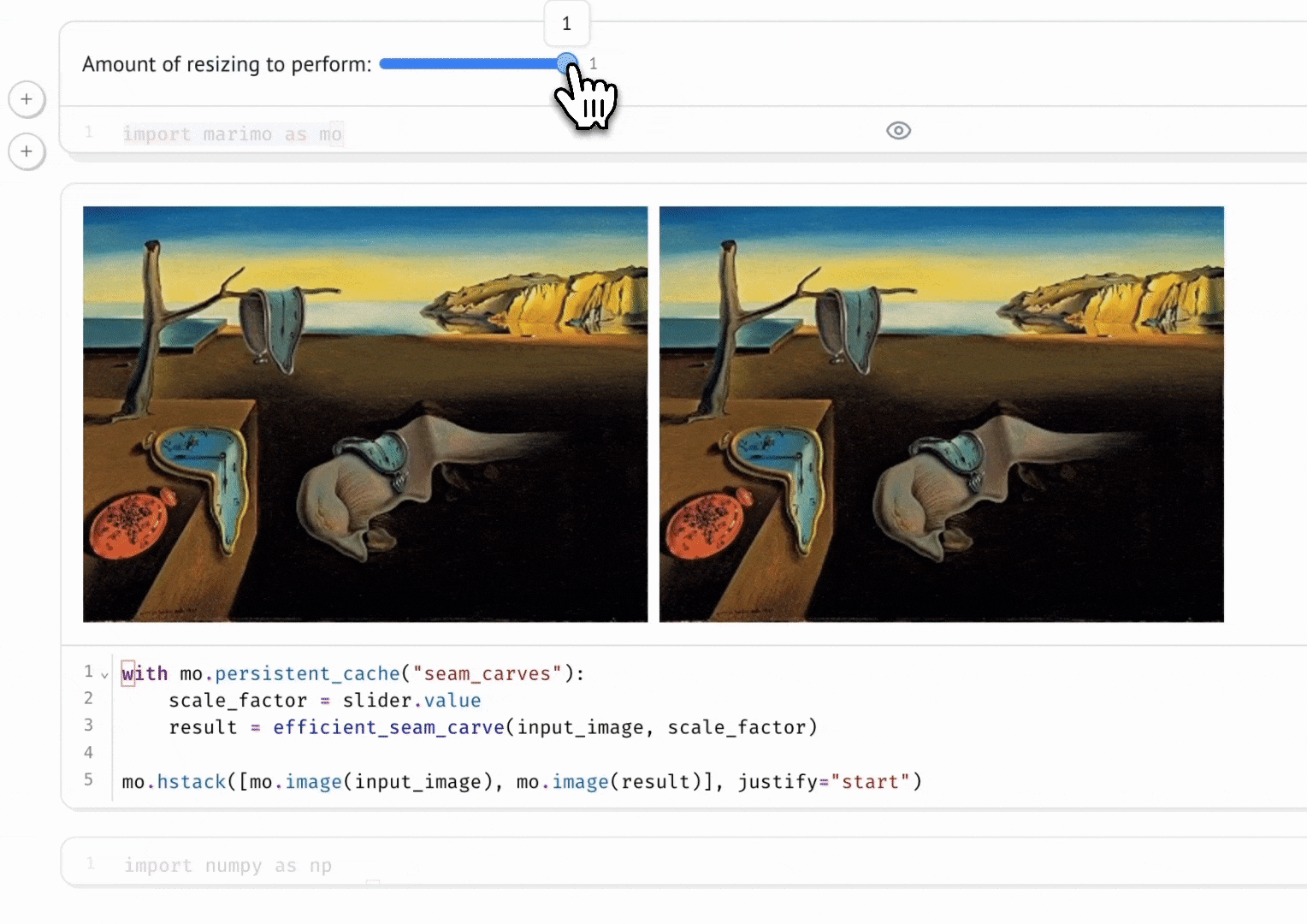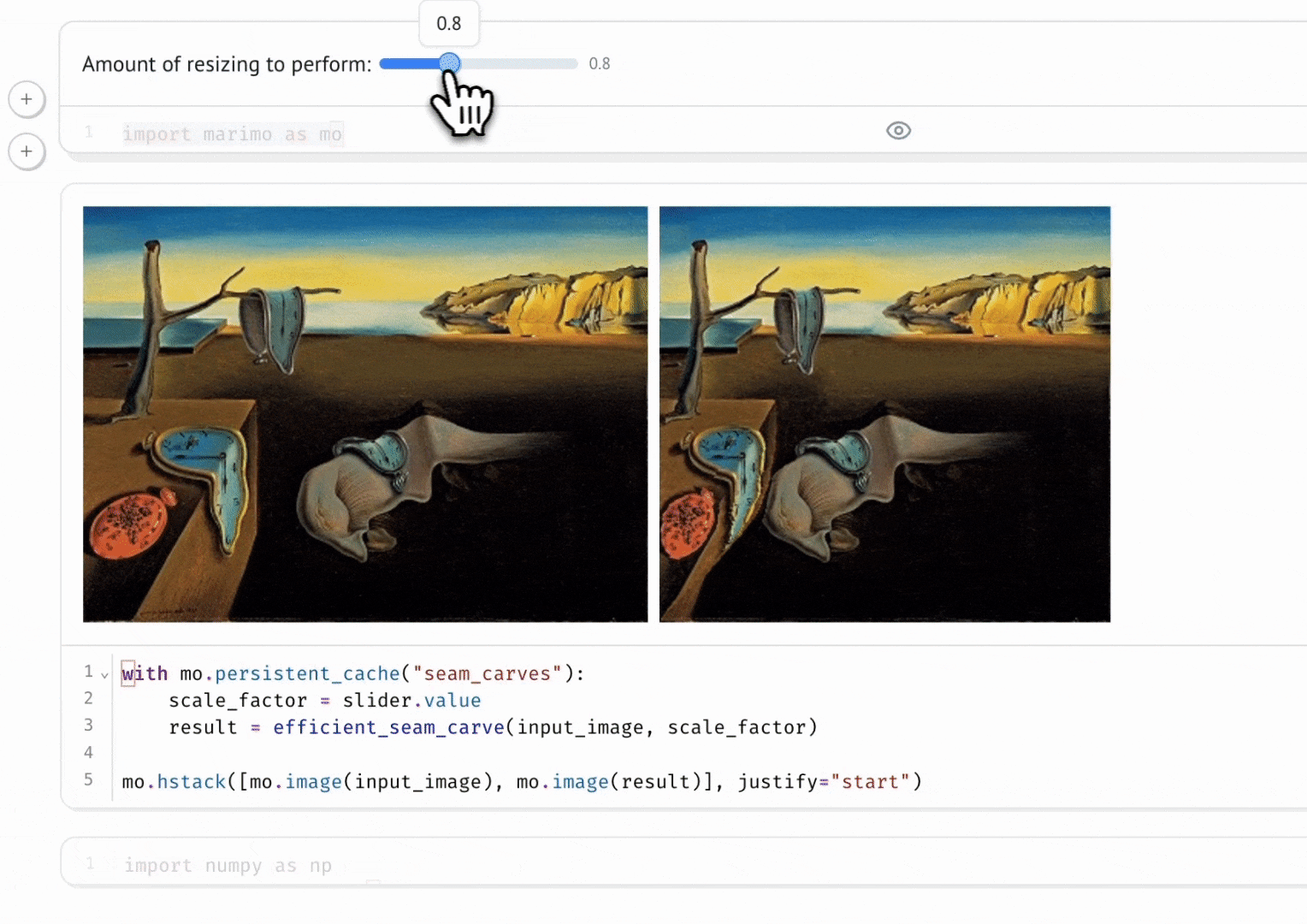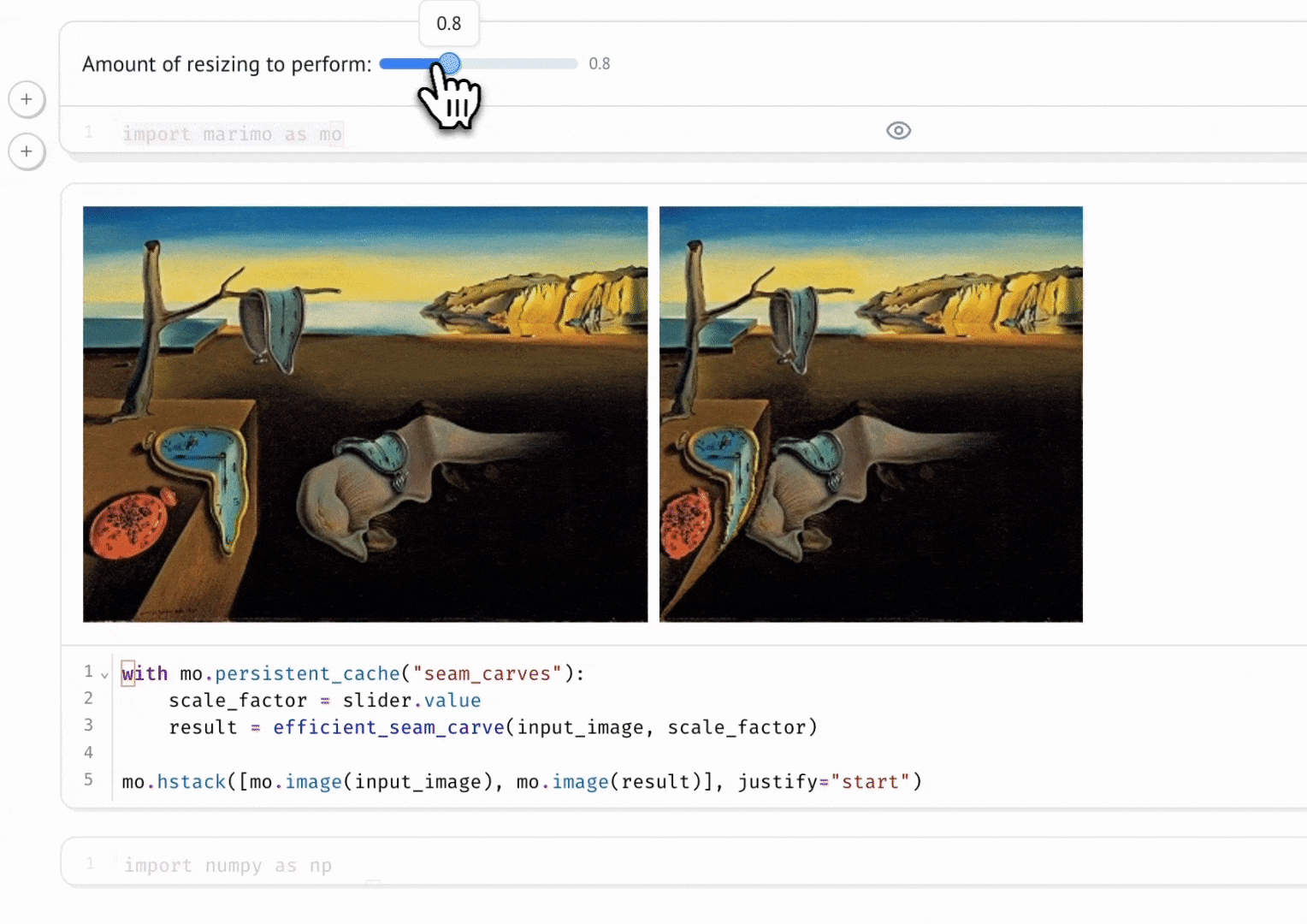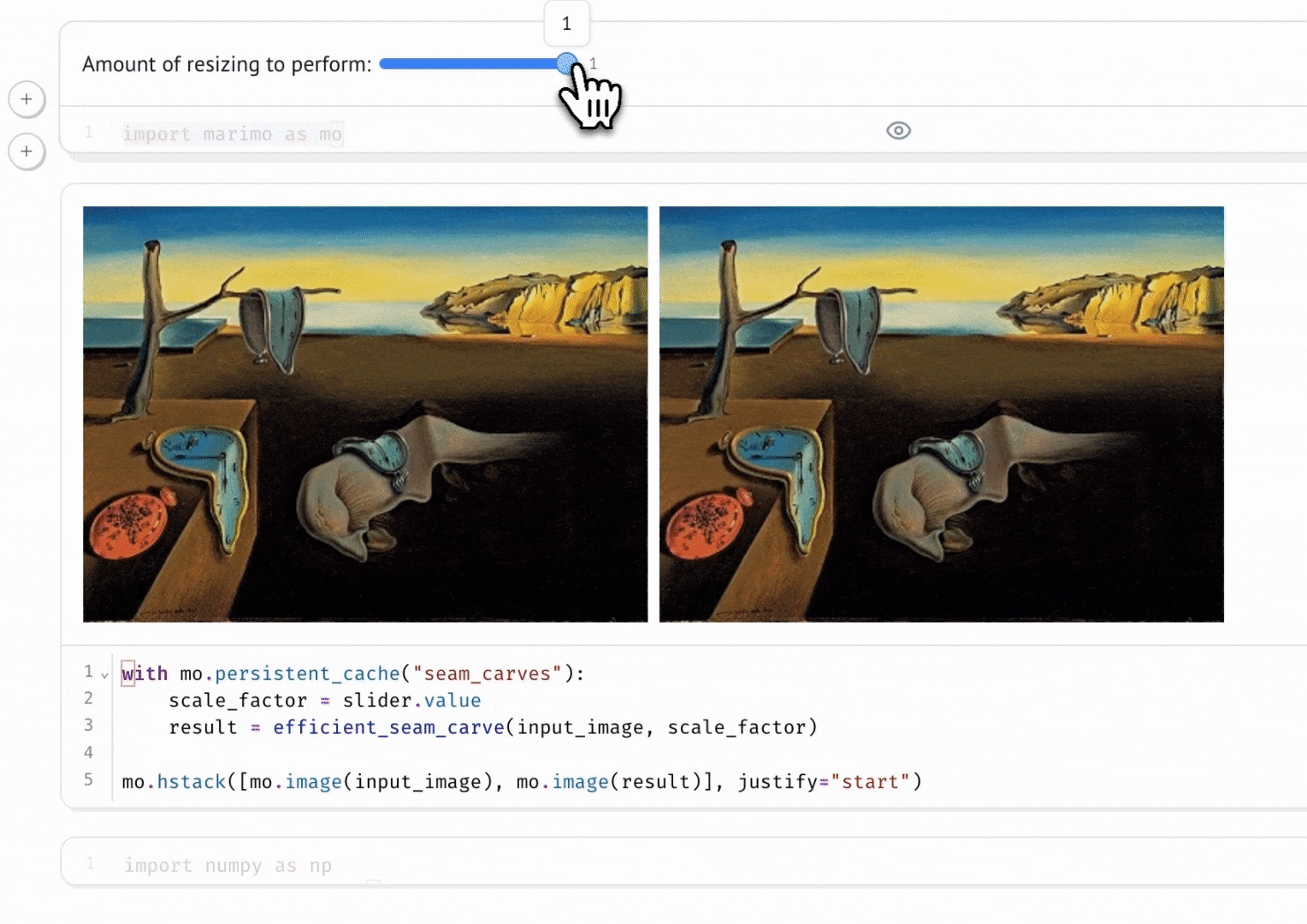
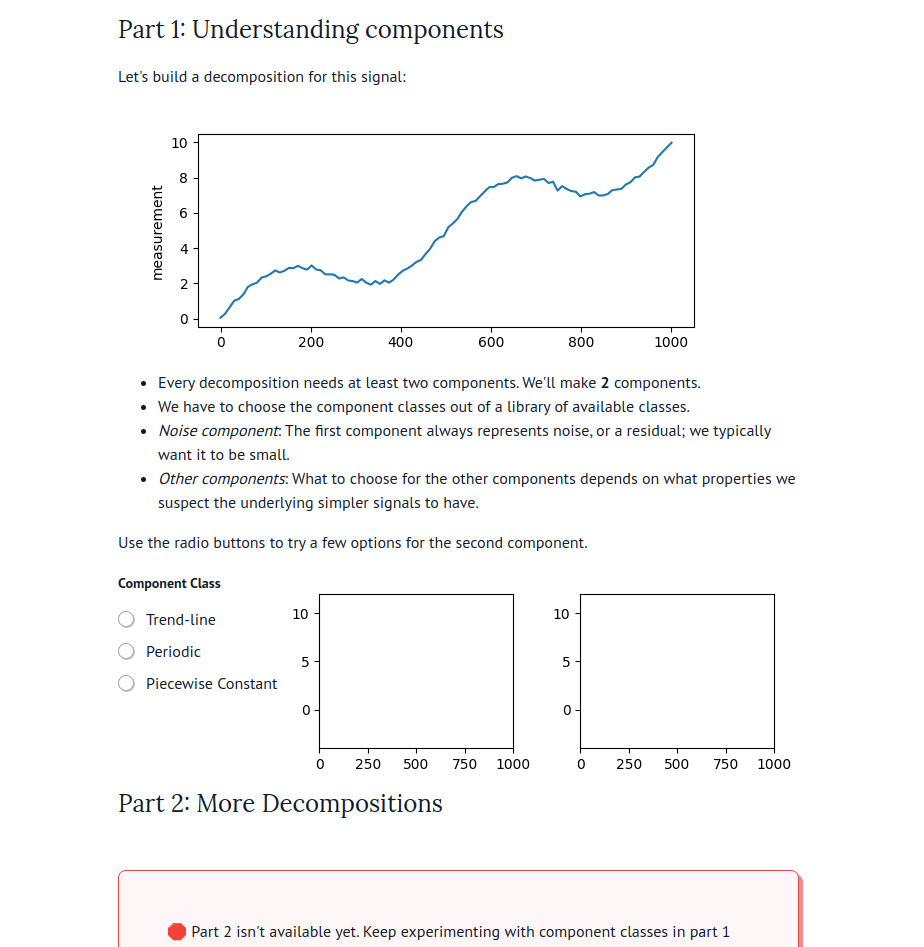
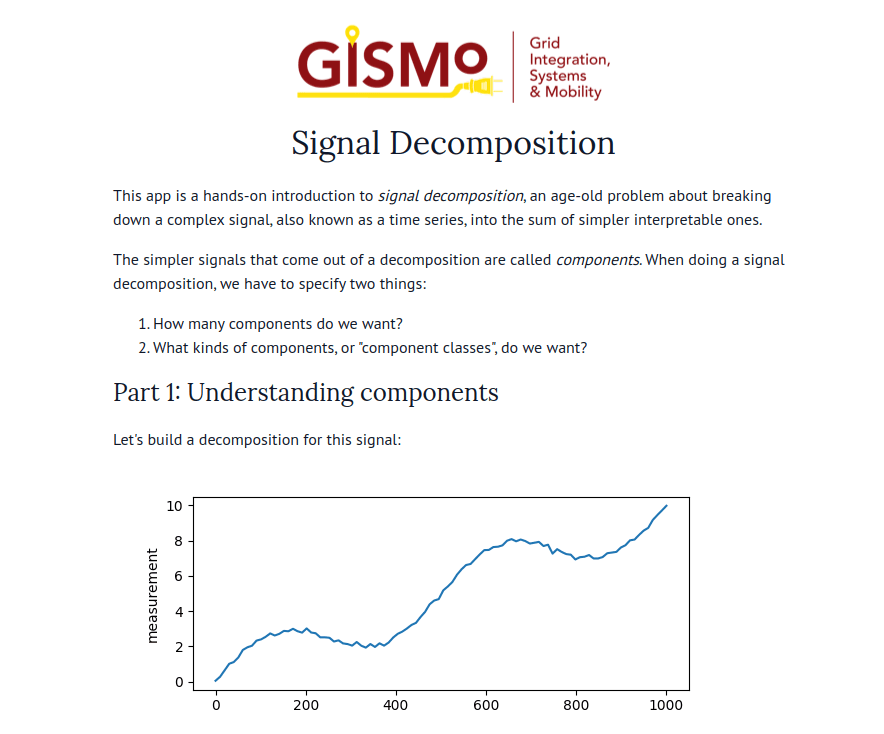
marimo’s combination of reactivity and interactivity is transformative for science communication: it allows scientists to publish interactive, computational articles on the web, without any training in web development, with just a single command. Indeed, GISMo’s first use case for marimo was exactly this: staff scientist Bennet Meyers used marimo to publish an interactive tutorial on signal decomposition methods for photovoltaic signals, allowing readers without a background in math or even coding to develop an understanding of advanced computational methods through a hands-on experience.
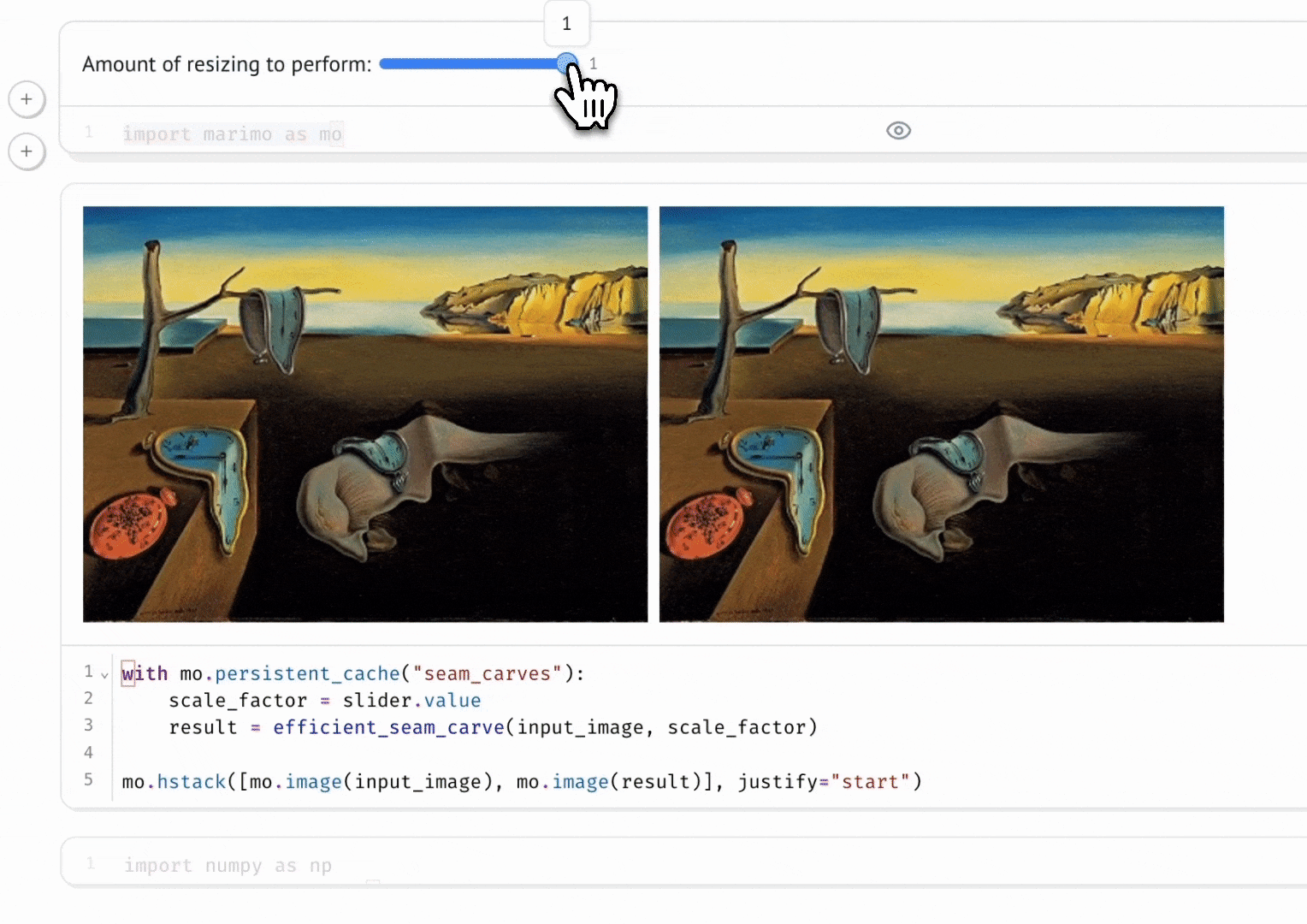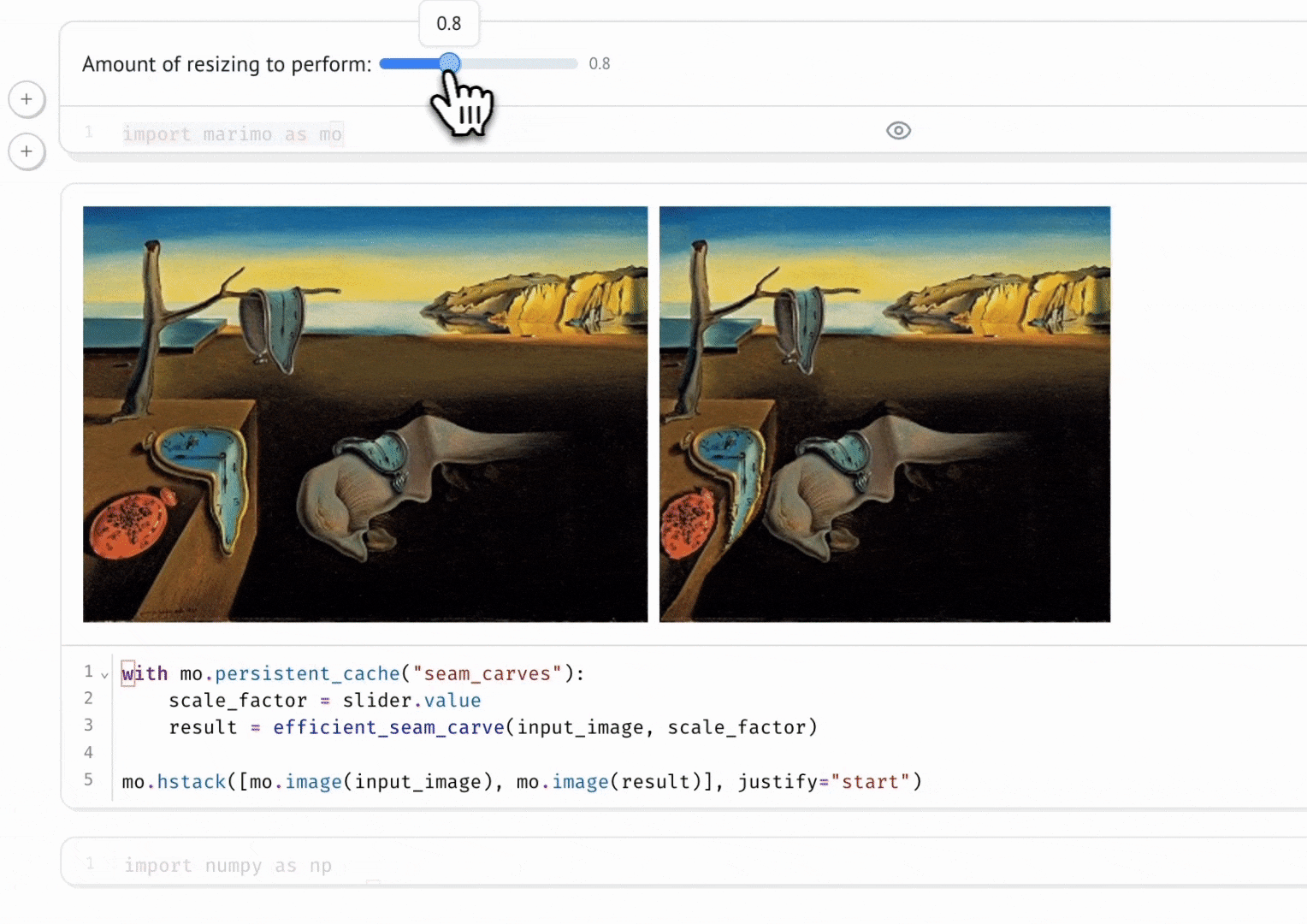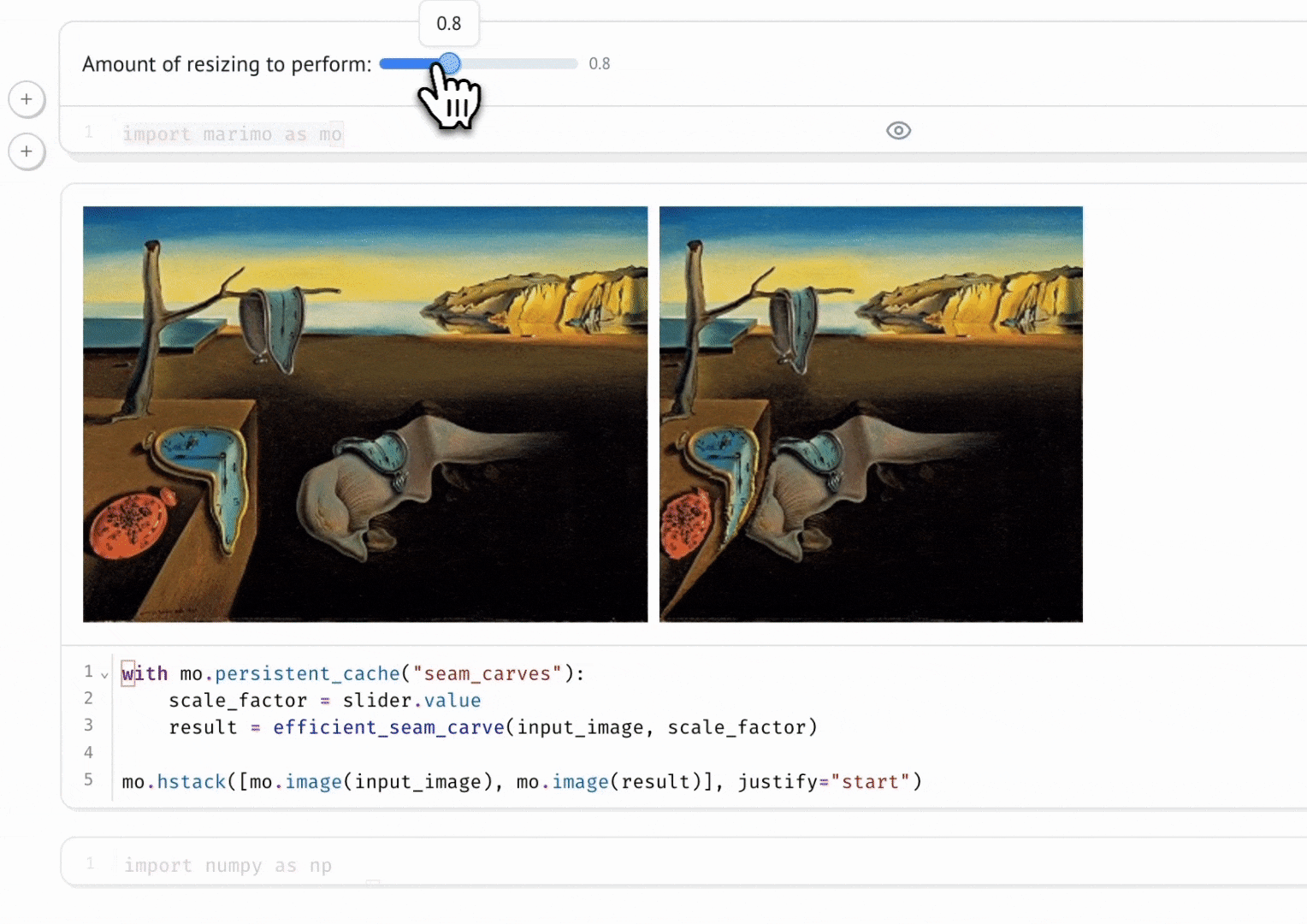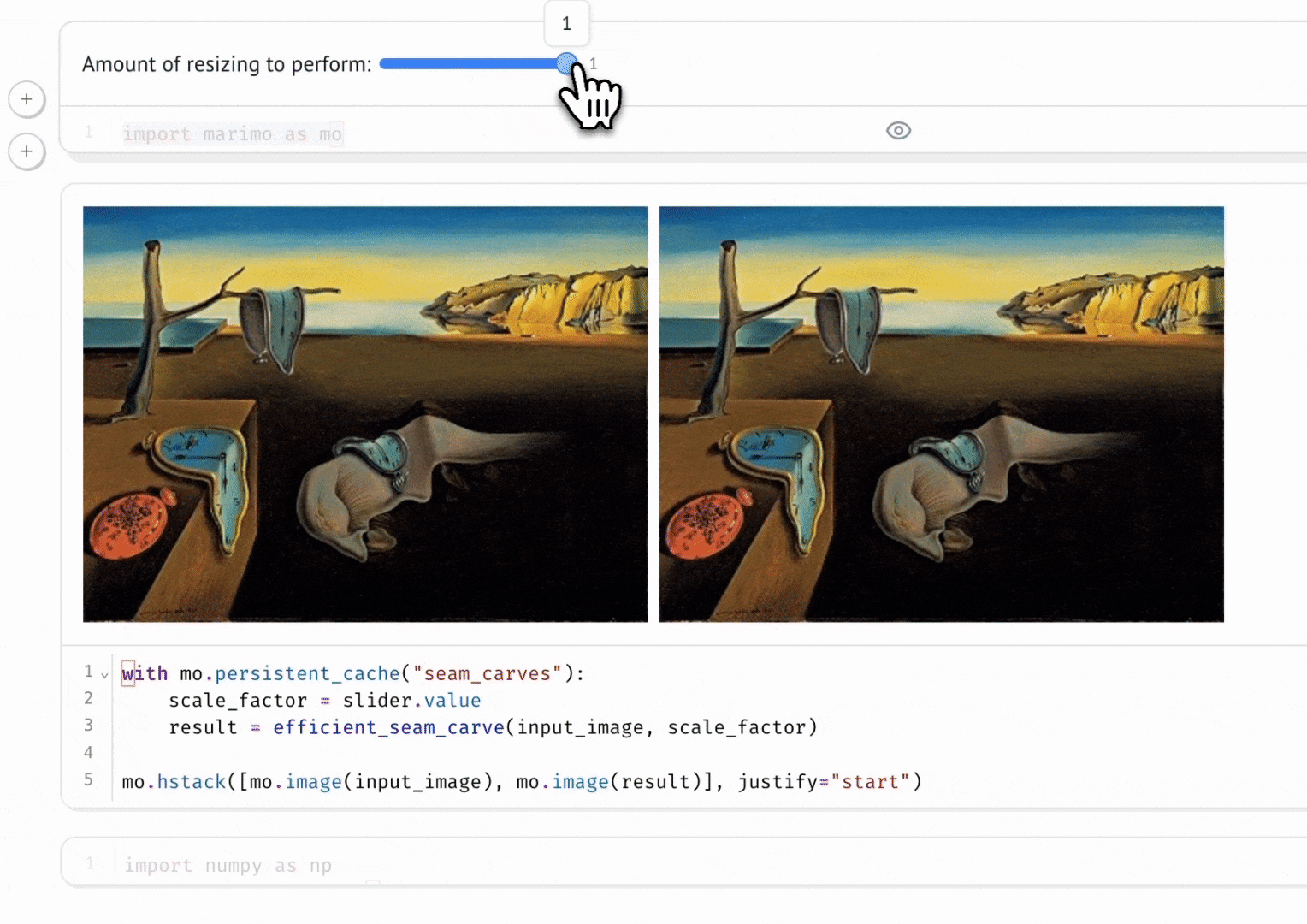
Seam Carving
Vincent Warmerdam
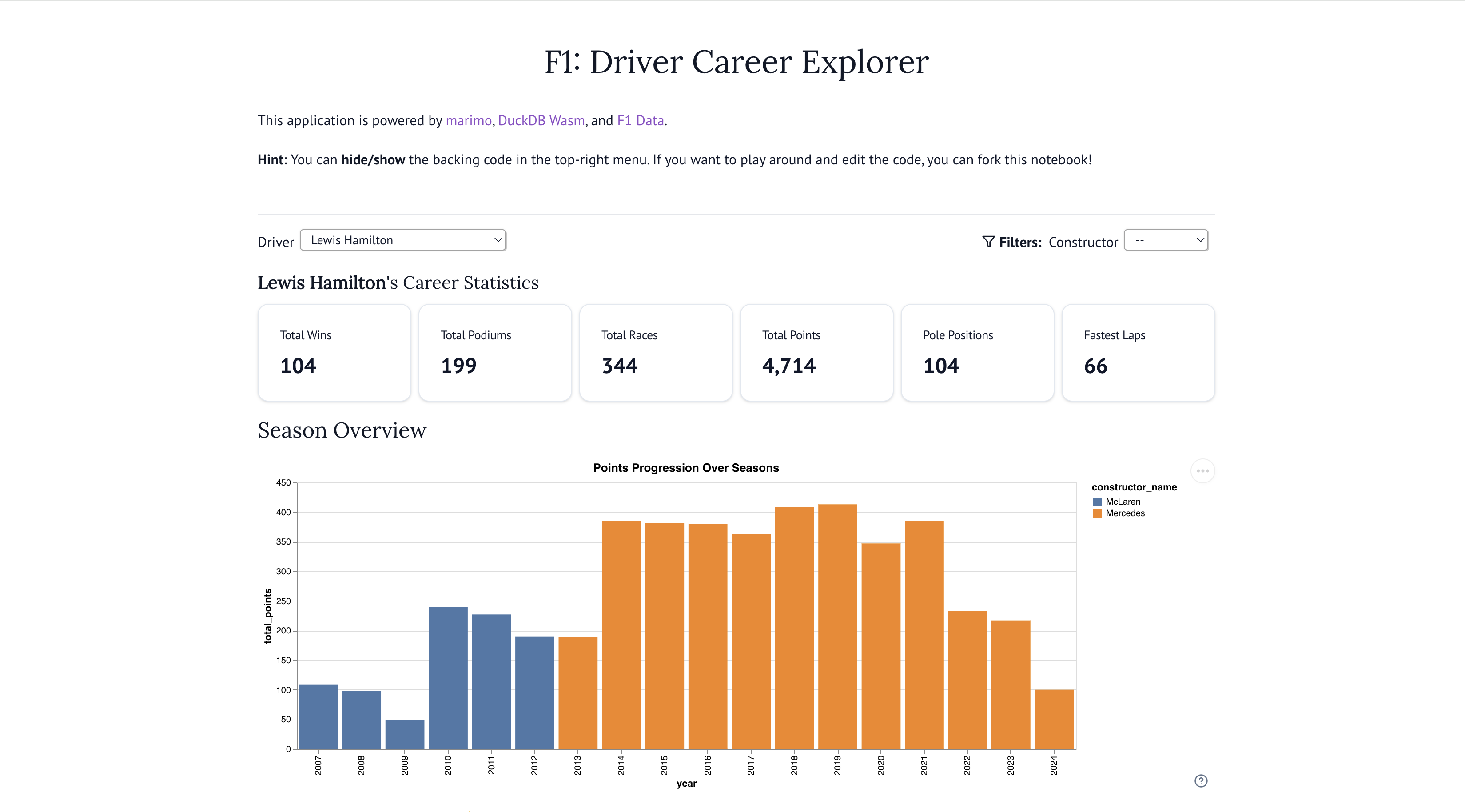
F1 Driver Explorer
marimo-team
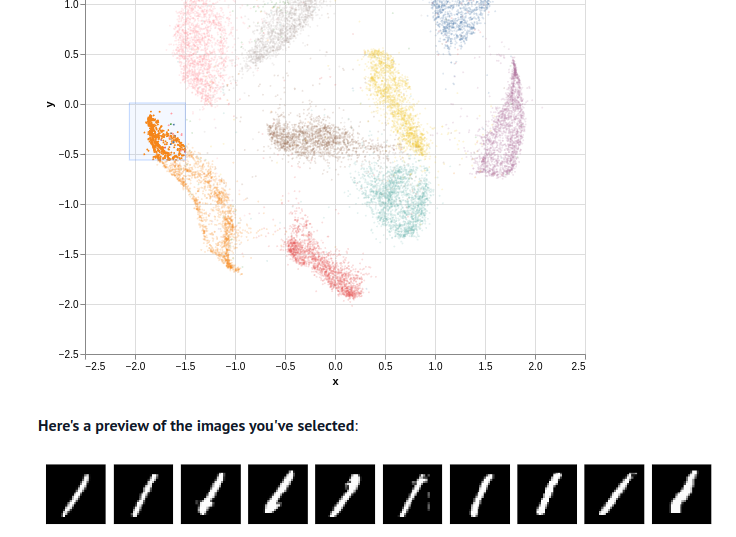
Embedding Visualizer
marimo-team
A gallery of interactive notebooks developed by the marimo community.
There are now dozens of educational marimo notebooks published to the web, including notebooks on convex optimization created for a course at NASA, a hands-on explainer of PCA, a primer on hypothesis testing by a Mozilla scientist, and many more. The interactive web apps produced by marimo need not be articles or tutorials; they can also be used as internal web-based tools. For example, David Chassin created a marimo notebook to control their lab’s electrical equipment.
Finally, interactive elements not only help with science communication; they also help in the scientific process, especially when exploring data or prototyping algorithms (“what happens if I tweak this parameter?”).
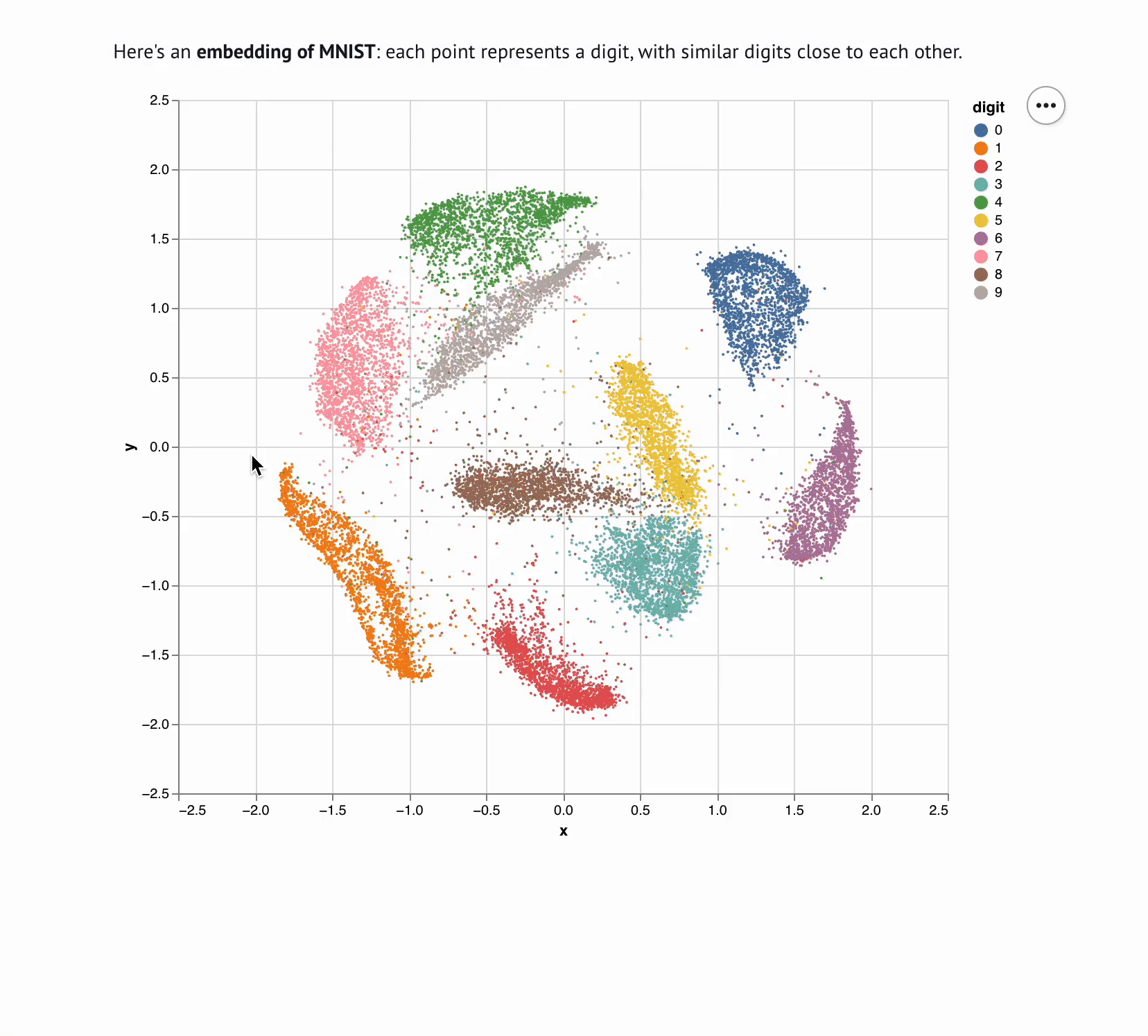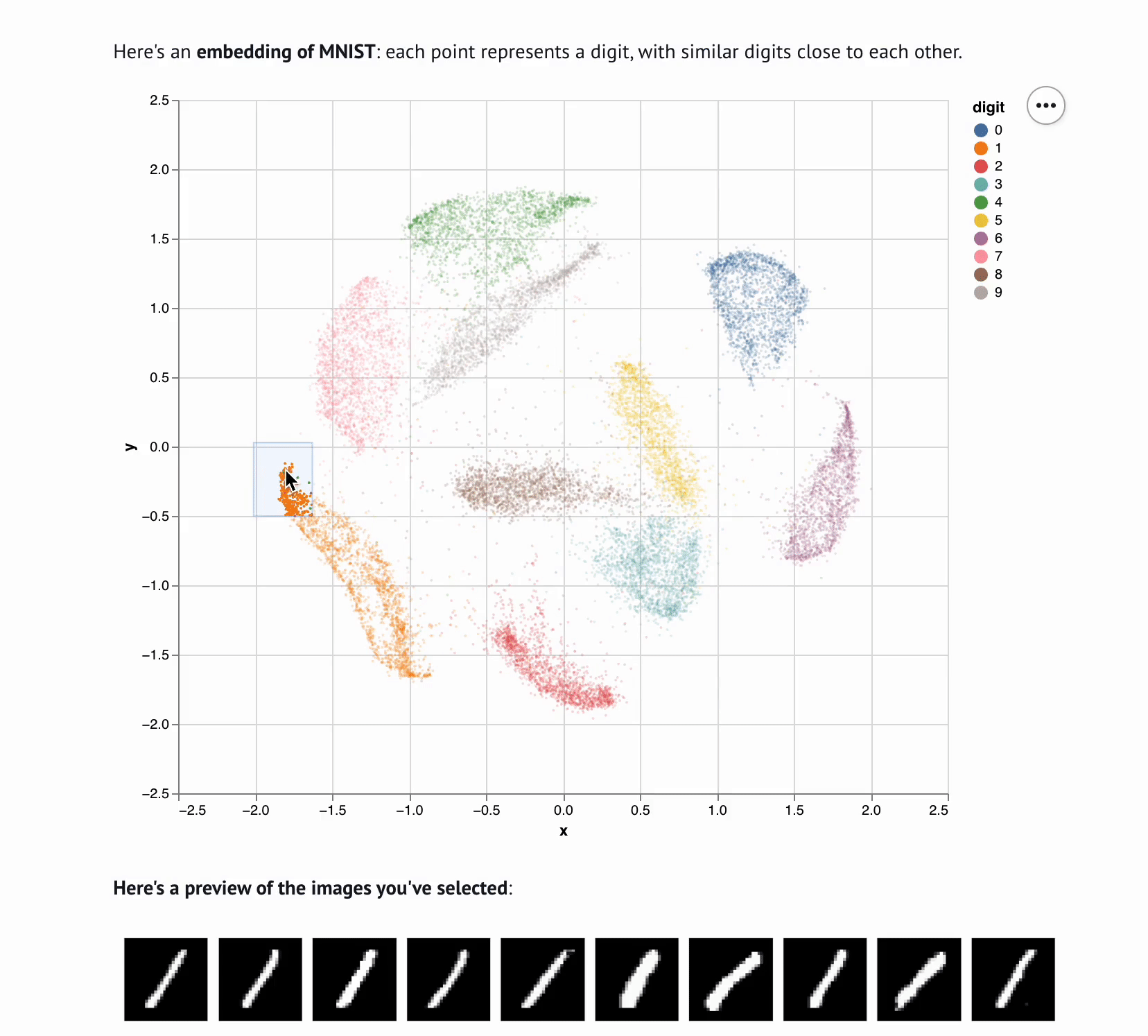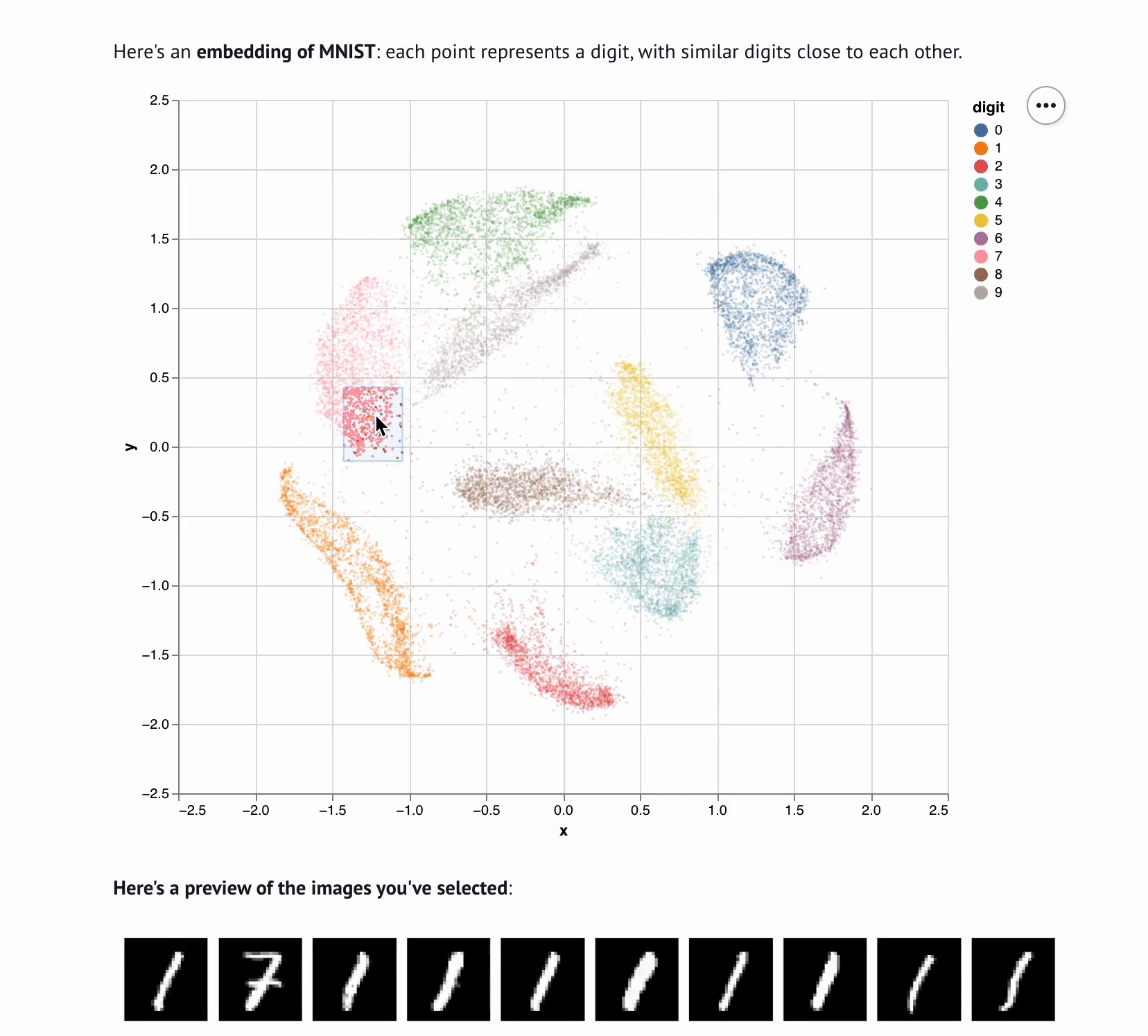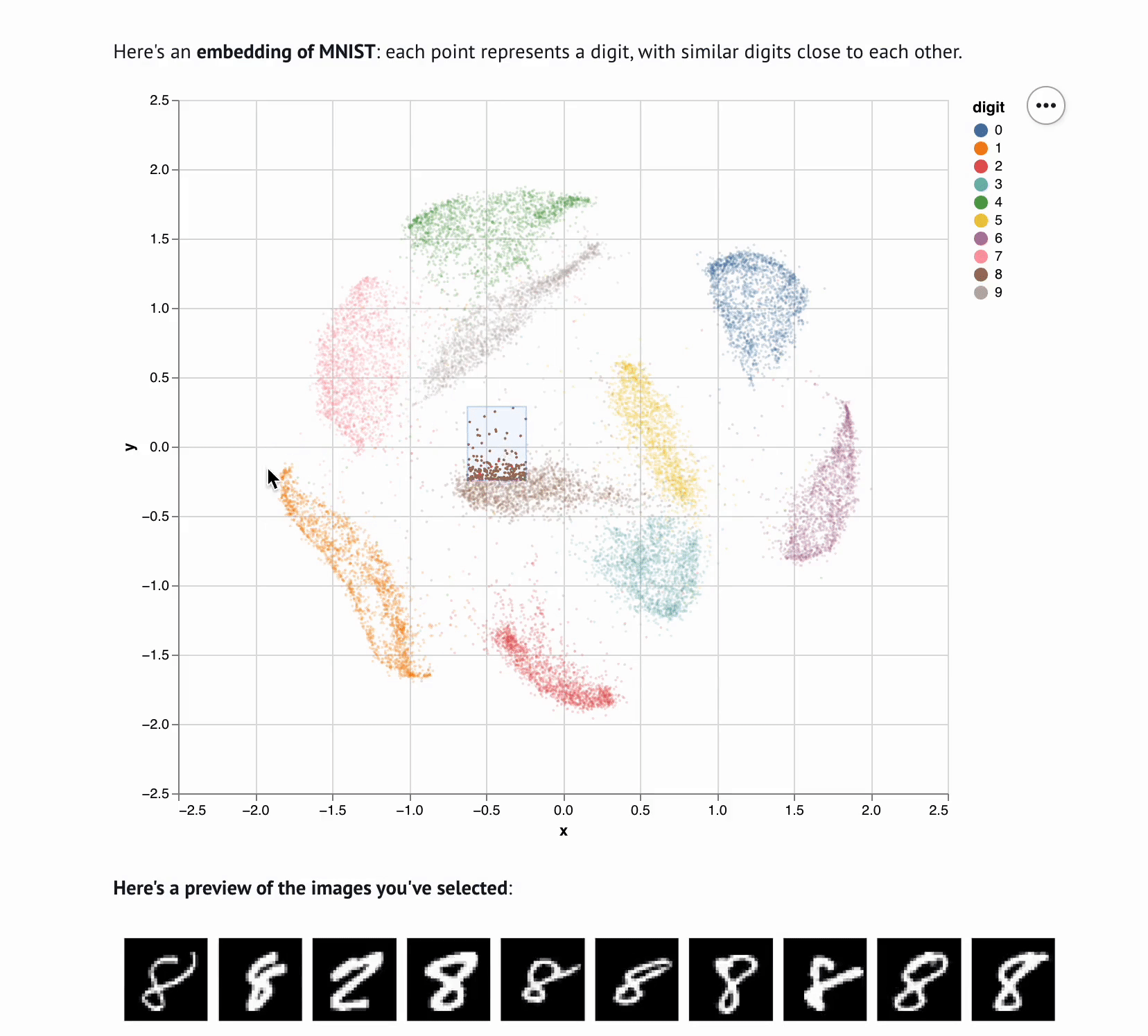
Sharing interactive notebooks without paying for compute
marimo notebooks can be published to the web without the complexity or cost of backend servers; instead, a technology called Pyodide executes the notebook’s code in the browser. The integration with Pyodide also makes it straightforward to embed marimo notebooks as part of projects’ online documentation, or even as part of interactive web-based textbooks.
Pyodide was originally developed by scientists at Mozilla as part of an experimental project designed for scientific communication and exploration on the web; in some sense, marimo is a realization of Pyodide’s original vision.
Providing instant feedback to Python learners
The immediate feedback reactive execution provides is well-suited to education.
GISMo hosted a group of undergraduate interns for a data science and research intensive in the summer of 2024, and had them use marimo for the duration of their internship. These students reported that marimo’s reactive execution made learning programming fun. As another example, Pluto.jl, a reactive notebook for Julia designed for education at MIT, powers the Introduction to Computational Thinking course at MIT.
A framework for reproducible computational experiments
Last year, the authors of this article collaborated to design a framework for running computational experiments driven by marimo notebooks.
Using the “expd” (experiment design) framework, scientists design and run a computational experiment by first implementing a “trial function” in Python, defining its inputs and outputs. Then, the expd framework uses a graphical user interface, implemented as a marimo notebook, to guide the scientist in designing an experiment — a collection of independent execution of the trial function with varying inputs generated by strategies that the scientist picks, such as gridding or random sampling.
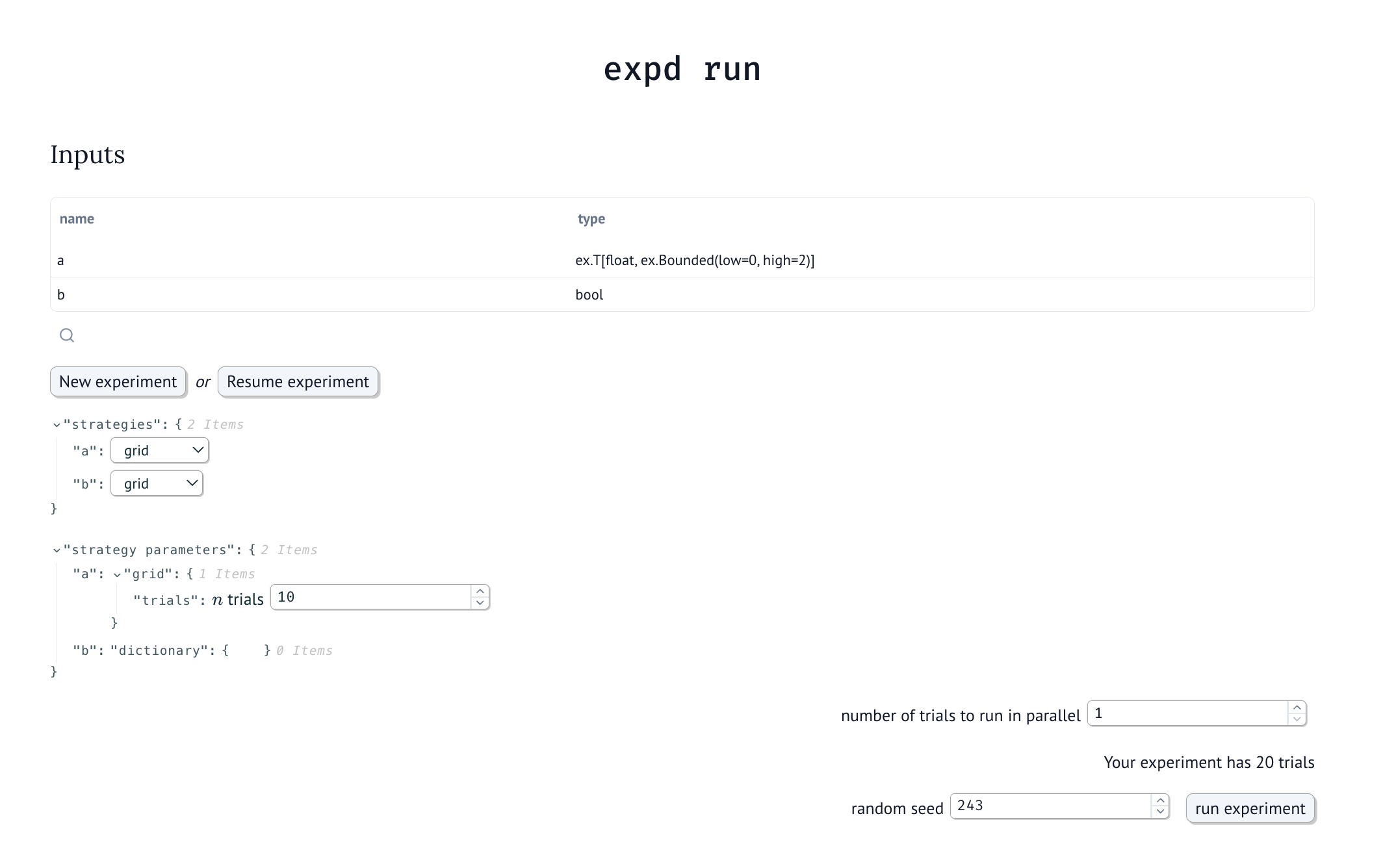
After selecting their strategies, the scientist simply clicks a button in the notebook to the experiment trials; the expd framework handles execution, visualizing results in a table, and saving them to disk. Interrupted experiments can be safely resumed, and the results of completed experiments can be viewed using another marimo notebook. Expd records the package and computational environment used to run the experiment, letting other scientists rerun it on their own machines.
The expd framework is just one example of the kinds of tools that marimo lets scientists build, and it is only possible because of the innovations that marimo brings to notebooks — reactivity, reproducibility, and resuability.
Enabling reproducibility in computational science
marimo belongs to a family of next-generation reactive notebooks that are different turns on the same perspective: that computational notebooks should be reactive, reproducible, and reusable programs, not error-prone scratchpads.
Since its release, marimo has been downloaded over a million times, and has received contributions from over 80 developers. While marimo’s roots are in computatational science, it is now used by dozens of companies and universities across the world, for a wide range of tasks.
We are hopeful that marimo will contribute to greater reproducibility and seamless communication of scientific research, and will one day enable a future where every computational research paper has an associated code artifact whose outputs are verifiably reproducible and easily shareable.
- GISMo: https://gismo.slac.stanford.edu/
- marimo: https://github.com/marimo-team/marimo
- Expd: https://github.com/marimo-team/expd
- Discord: https://marimo.io/discord
- Docs: https://docs.marimo.io
- Newsletter: https://marimo.io/newsletter