For researchers
marimo is an open-source reactive Python notebook designed specifically for reproducible computation, rapid data manipulation, and interactive science communication on the web. It solves long-standing problems with traditional Python notebooks while also giving researchers new capabilities:
- Reproducible execution and packaging. Run a cell and marimo runs affected cells (or marks them as stale), eliminating hidden state and hidden bugs that affect traditional notebooks.
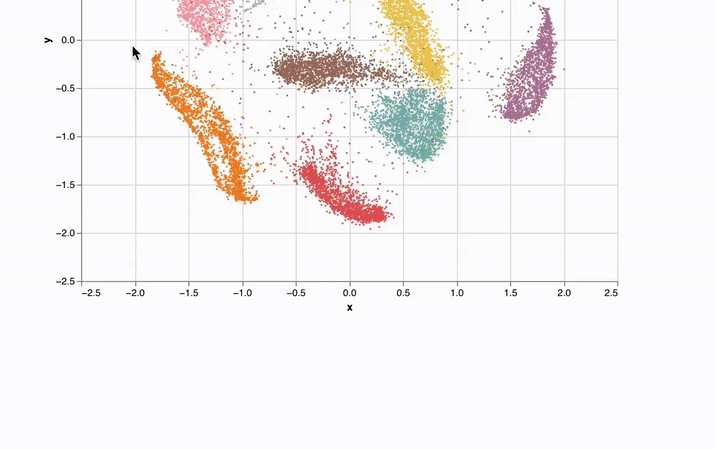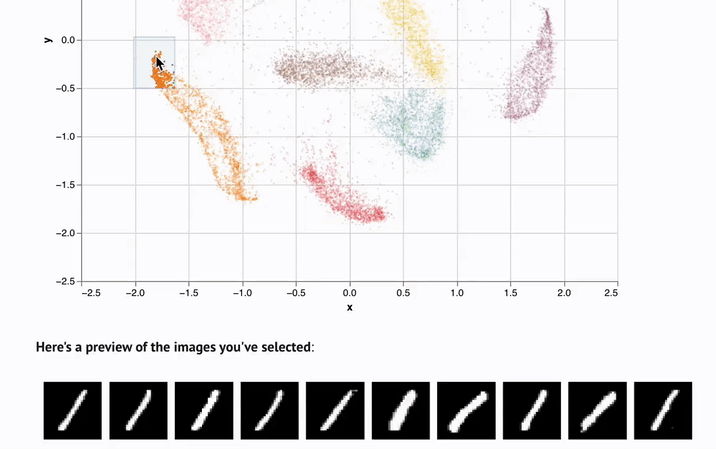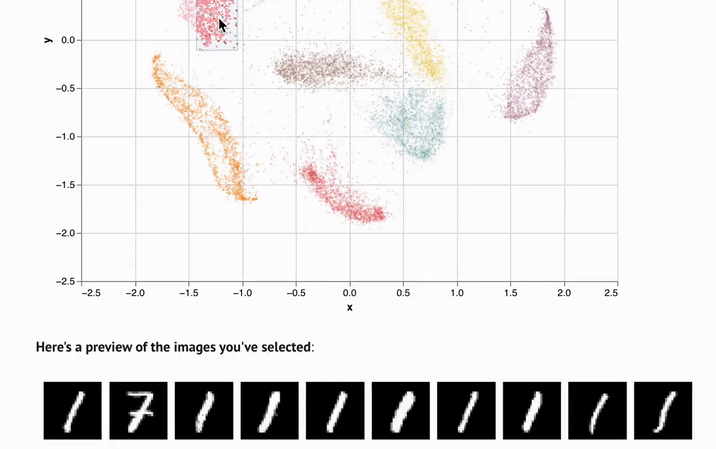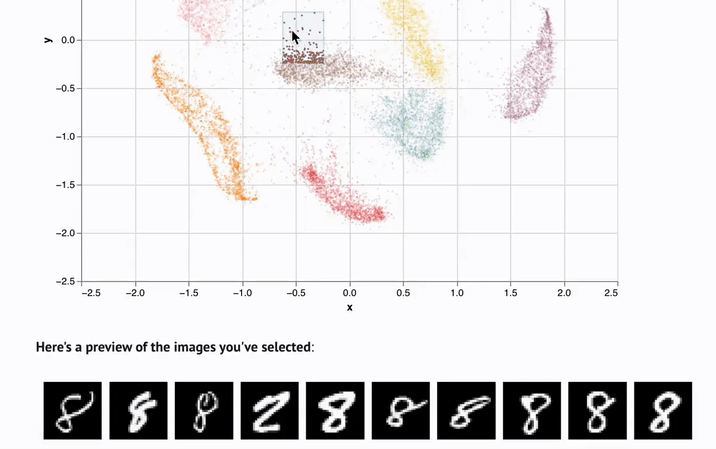
- Interactivity. UI elements like sliders, scatterplots, and infinitely scalable tables are automatically synchronized with Python, letting you experiment with data in new ways.
- Pure Python file format. A Python file format makes it easy to version with Git, run as scripts, or reuse as modules.
- Share as web apps. Every notebook optionally doubles as an interactive web app that can be published on GitHub Pages or deployed on a server.
Minimum-distortion embedding
Akshay Agrawal | Stanford
Agentic Visual Reporting
Péter Gyarmati | Vienna, CMU

Signal Decomposition
Bennet Meyers | Stanford
marimo was originally designed by Stanford scientists, for Stanford scientists. Today, marimo is used at companies, labs, and classes around the world, has been downloaded millions of times, and runs everywhere: locally, in VS Code, in the cloud, and even inside web pages like this one. Try marimo below!
Getting started
Here are some resources to help you get started with marimo:
- Run locally. marimo is free and open source; use the command-line interface or VS Code.
- Run on the cloud. Use molab, our free cloud-hosted notebook; no setup required.
- Primer for education. See our self-contained primer on marimo for educators and students.
- Classes using marimo. See this list.
- Examples. See our notebook gallery.
- Documentation. See our online documentation.
- Videos. Our YouTube has many tutorials.
- Forum. Join our discussion forum to chat with the marimo team and other researchers.
- Feature requests or issues. Reach out on GitHub with feature requests or issues.
Features
Reproducible by default
Traditional notebooks suffer from a reproducibility crisis. One study found that less than a quarter of Jupyter notebooks were executable, and less than four percent of those were reproducible. The default experience in Jupyter notebooks accumulates hidden state: run a cell and the kernel imperatively mutates memory; the user must remember to explicitly run affected cells, a tedious and error-prone process. While there are steps the practiced scientist can take to make Jupyter-driven work reproducible, they require awareness of Jupyter's many pitfalls as well as the know-how and discipline to steer clear of them.
In contrast, marimo notebooks are"reproducible by default" (a phrase we borrow from Pluto.jl). Run a cell and marimo reactively runs affected cells based on a statically parsed dataflow graph (for expensive notebooks, users can disable automatic execution, with marimo marking affected cells as stale instead of automatically running them). Install a package, and marimo can not only serialize dependencies in your notebook file, but also create ephemeral, isolated virtual environments, making your notebooks reproducible down to the packages.
To learn more, see the marimo feature published in Nature.
Interactivity with sliders, scatter plots, and more
marimo notebooks come packaged with UI elements, which are automatically bound to Python code. Make a selection on a scatterplot, and marimo automatically sends the selected points back to Python as a dataframe, making data tangible in a way that it just isn't in traditional notebooks.

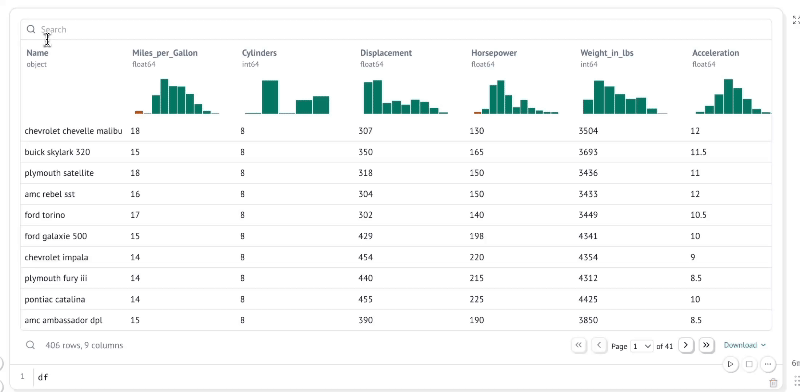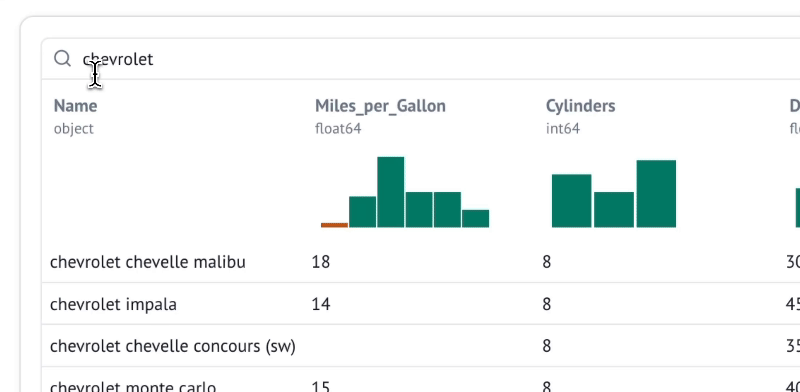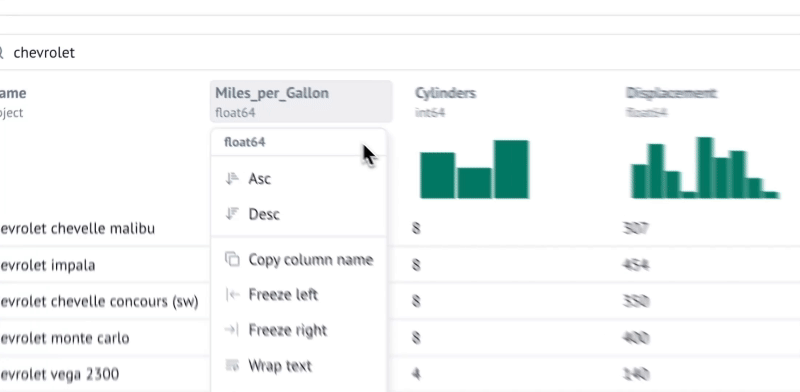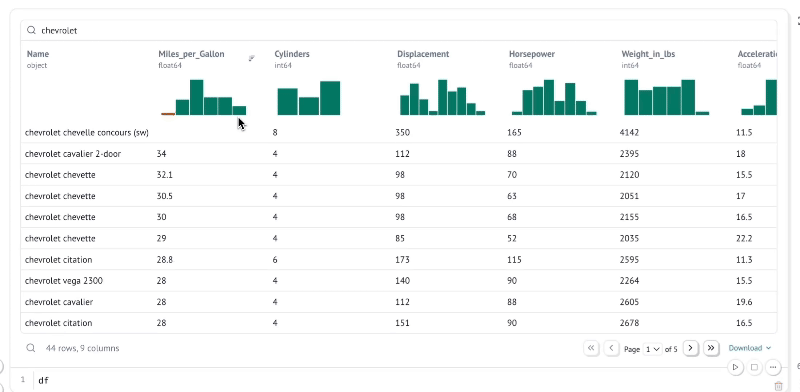
Explore data with infinitely scalable tables
marimo's built-in dataframe viewer lets you search, sort, filter, and page through as much data as fits in your machine's RAM, which on modern machines could easily be terabytes.
Create custom widgets with anywidget
If you know "just enough JavaScript", you can extend marimo with custom UI elements or "widgets", like the matrix widget below.
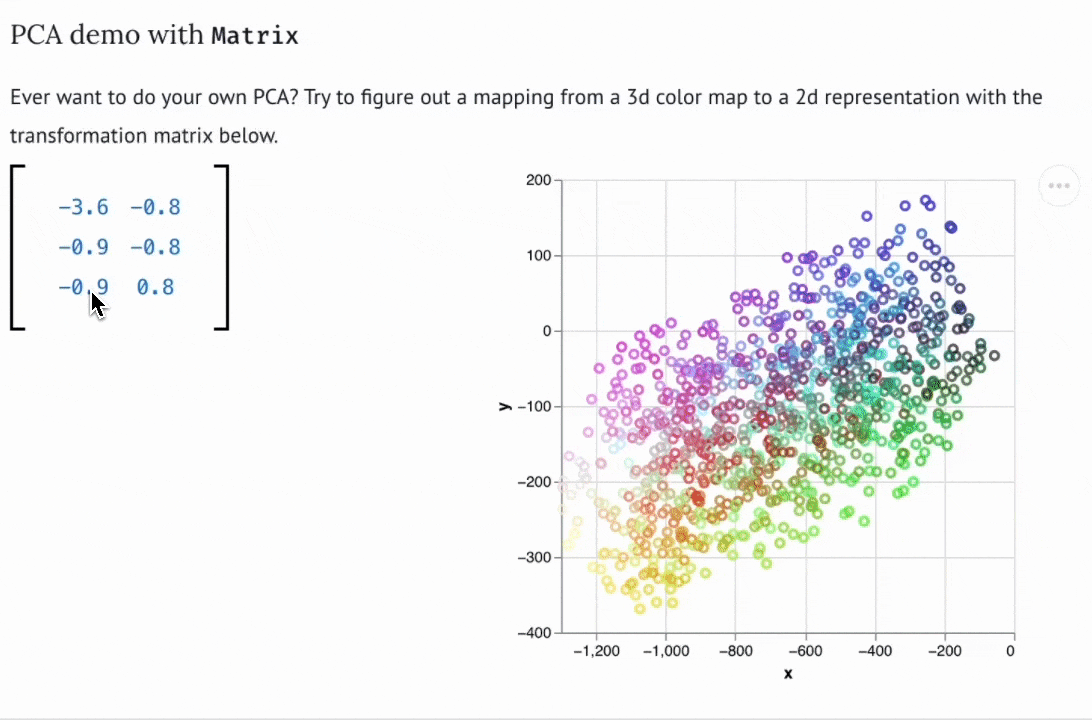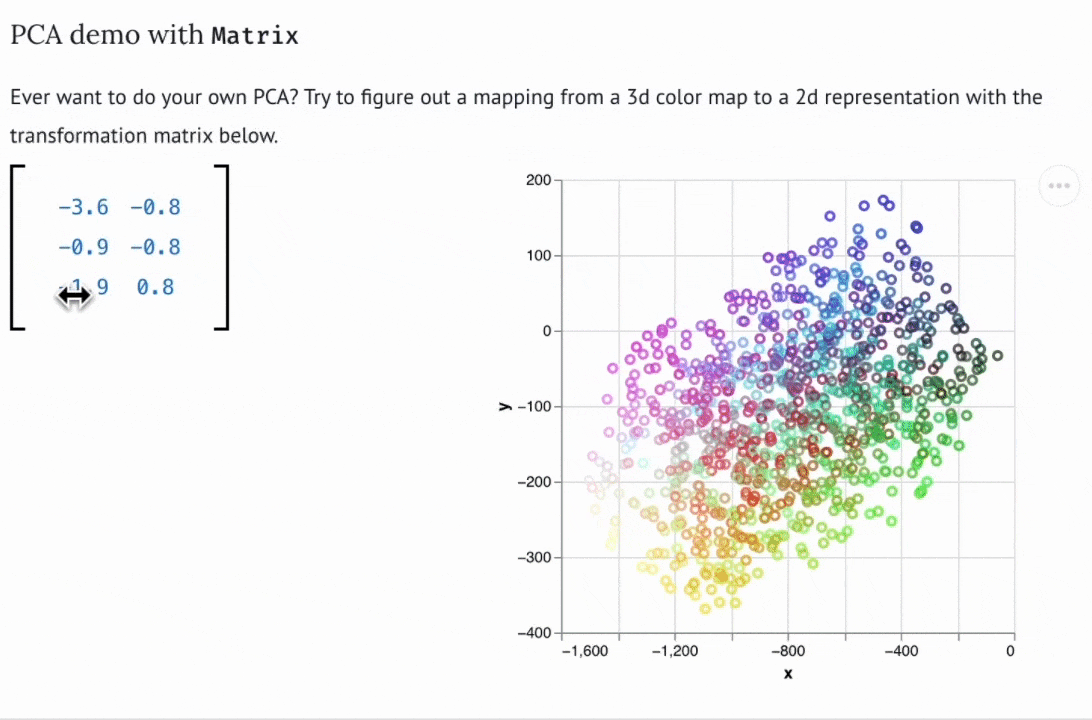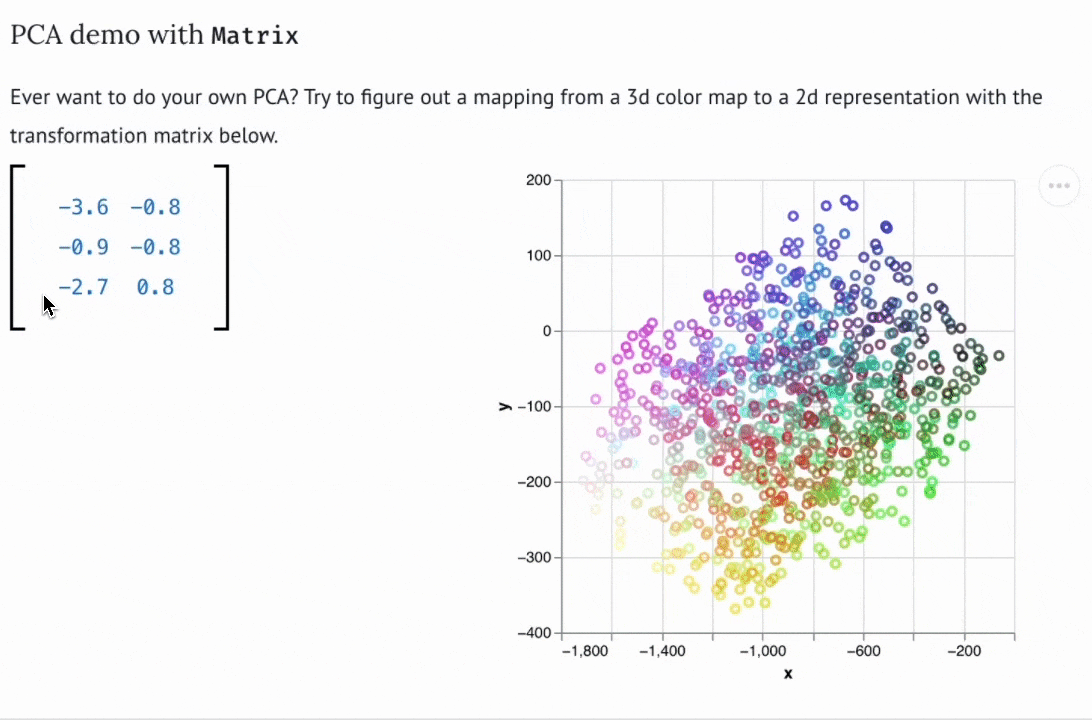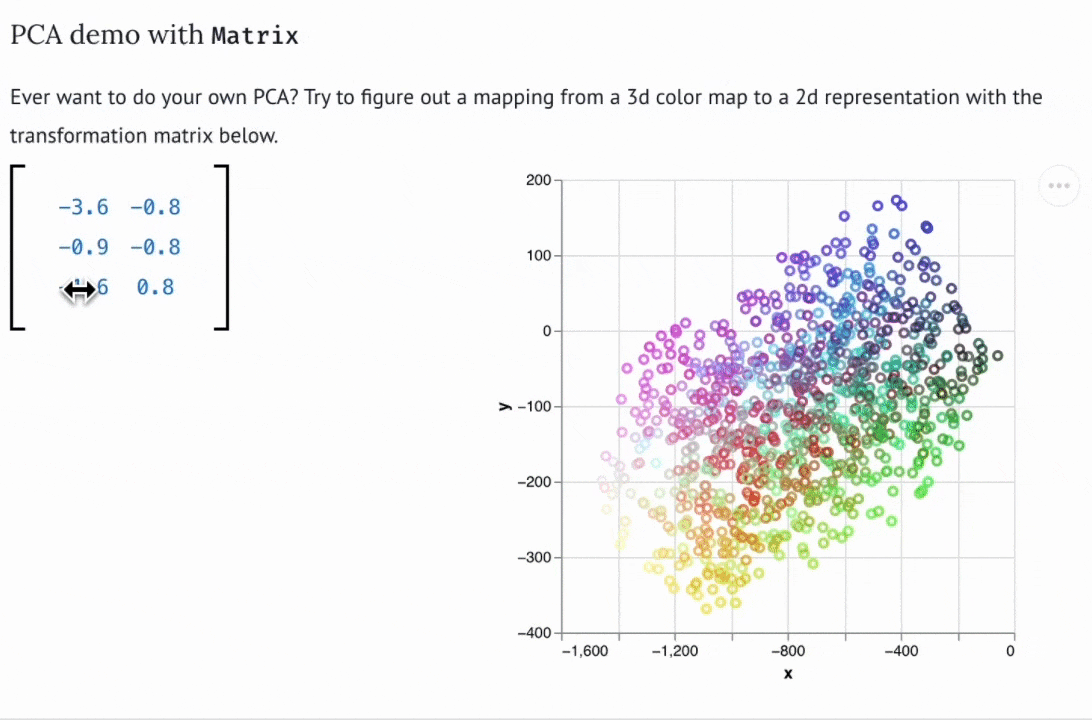
You can use bespoke widgets to accelerate domain-specific workflows such as data labeling or geospatial data analysis. If you don't know JavaScript, you can still benefit from marimo's ecosystem of third-party widgets, which is compatible with anywidget. Check out the wigglystuff project for many examples of custom widgets (including the Matrix widget).
Easily share on the web
Researchers can share marimo notebooks on the web in one of many ways:
- export to ipynb or HTML;
- embed in web pages;
- link to notebooks hosted on GitHub;
- publish on molab, our free cloud hosted service;
- deploy on a server, using our CLI and your own backend.
Use software engineering best practices
marimo sets you up for success for industry or academia by gently encouraging software engineering best practices: marimo notebooks are stored as pure Python, so they can be version controlled with Git; they are testable with PyTest; and they encourage functional-style programming with minimal mutations.